Abstract
Failure to eliminate abnormal proteins in the cell is associated with numerous aggregation diseases. Misfolded proteins are normally detected by protein quality control and either refolded or eliminated. The ubiquitin-proteasome system is a major pathway that degrades these unwanted proteins. Ubiquitin ligases are central to these degradation pathways as they recognize aberrant proteins and covalently attach a polyubiquitin chain to target them to the proteasome. We discovered that the Hul5 ubiquitin ligase is a major player in a novel protein quality control pathway that targets cytosolic misfolded proteins. Hul5 is required for the maintenance of cell fitness and the increased ubiquitination of low solubility proteins after heat-shock in yeast cells. We identified several low-solubility substrates of Hul5, including the prion-like protein Pin3. It is now apparent that in the cytoplasm, misfolded proteins can be targeted by multiple degradation pathways. In this review, we discuss how the Hul5 protein quality control pathway may specifically target low solubility cytosolic proteins in the cell.
Ridding of the Unwanted Proteins by Protein Quality Control
Protein misfolding and aggregation can threaten cell viability and are linked to multiple diseases with a strong prevalence in age-related neurodegenerative pathologies, such as Parkinson and Huntington’s. To prevent the cytotoxic accumulation of misfolded proteins, the cell has developed protein quality control (PQC) pathways that rely on networks of molecular chaperones and proteolytic machineries.Citation1,Citation2 PQC can either assist misfolded proteins to refold, or eliminate the ones that have failed to achieve or maintain their native structures. These misfolded polypeptides are largely degraded by the ubiquitin proteasome system. A critical challenge for the cell is to distinguish between transiently and terminally misfolded proteins and only target the latter for proteolysis, a harrowing task due to the large variability of protein structures and folding rates. Comprehending how PQC separates “the good from the bad” is the key to unearthing the underlying mechanisms leading to disease, and therefore the development of novel therapeutics.Citation3
One line of defense for the cell to combat misfolding is to adapt distinct strategies based on the localization of the misfolded proteins. For example, endoplasmic reticulum (ER)-localized misfolded proteins are degraded by ER-associated protein degradation (ERAD) pathway.Citation4 In this pathway, misfolded proteins first undergo retro-translocation to the cytoplasm, followed by the covalent attachment of poly-ubiquitin chains for proteasome degradation. Polyubiquitination requires a series of enzymes: ubiquitin activating enzyme (E1); ubiquitin conjugating enzyme (E2); and ubiquitin ligase (E3).Citation5 As ubiquitin ligases mediate substrate recognition, they are the “signature components” of the different PQC degradation pathways. In yeast, ER misfolded proteins are targeted by the conserved Hrd1 and Doa10 ERAD ubiquitin ligases,Citation4 while aberrant nuclear proteins are ubiquitinated by the San1 ubiquitin ligase.Citation6
It is becoming apparent that in the cytoplasm-the epicenter of protein synthesis and folding- the cell can deploy a variety of different degradative PQC pathways. The CHIP (carboxy terminus of Hsp70-interacting protein) is a major PQC ubiquitin ligase in metazoans that selectively ubiquitinates chaperone-targeted misfolded proteins for proteasomal degradation.Citation7,Citation8 CHIP, which is not conserved in lower eukaryotes like yeast, is not the sole E3 that targets cytosolic misfolded proteins to the proteasome. For instance, the N-end rule Ubr1 ubiquitin ligase not only targets polypeptides with destabilizing N-terminal amino acids (e.g., arginine) for degradation, but also mediates chaperone-dependent ubiquitination of cytosolic proteins that are internally misfolded.Citation9,Citation10 In mammalian cells, the Ubr1 and CHIP pathways are redundant (at least for a subset of misfolded proteins).Citation11 In yeast, Ubr1 can function together with the Ubr2 or the nuclear-localized San1 ubiquitin ligases.Citation9,Citation10,Citation12 Another PQC pathway in the cytoplasm requires the ubiquitin ligase Rkr1/Ltn1 that directly associates with ribosomes to specifically target aberrant newly synthesized polypeptides that contain a translated poly(A) tail due to the absence or missed-stop codon.Citation13
We hypothesized that additional PQC pathways may target misfolded cytosolic proteins. The yeast and human genomes are estimated to encode 90 and 600 ubiquitin ligases, respectively, with most remaining uncharacterized. Thus, to uncover novel players of the cytosolic PQC, we characterized the heat-shock induced ubiquitination response and identified a novel pathway dependent on Hul5 (HECT ubiquitin ligase 5),Citation14 which is the focus of this Extra View.
Heat-shock Leads to the Ubiquitination of Cytosolic Proteins
Knowledge of yeast PQC is mainly based on the studies of model substrates that may not fully encapsulate the whole spectrum of physiological substrates. We reasoned that cellular stresses, such as transient and acute heat-stress that causes protein misfolding, could be used as a complementary approach to further study PQC. Indeed, we found that heat-shock stress triggers a rapid increase of poly-ubiquitination of low solubility proteins in yeast cells. Because the increase in ubiquitination occurs within minutes, this pathway likely requires constitutively expressed components that are present in the cell prior to the stress (as opposed to induced genes). Using quantitative mass spectrometry, we discovered that heat-shock primarily induces the ubiquitination of cytosolic proteins, including New1 and Pin3/Lsb2. These two proteins can induce [PSI+] prion when overexpressed.Citation15 Why are the majority of proteins ubiquitinated after heat-shock located in the cytoplasm? One possibility is that the ubiquitination of ER proteins, which have to be first retrotranslocated in the cytoplasm, is a slower process. As well, it was shown that heat-shock induces increased sumoylation (another ubiquitin-like post-translation modification on lysine residues) mainly of nuclear proteins in plant cells, as well as in mammalian cells.Citation16,Citation17 Therefore, sumoylation may prevent the ubiquitination of misfolded proteins in the nucleus by occupying the exposed lysine residues. Regardless of the exact mechanism, our results indicate that there are PQC components that mainly target cytosolic misfolded proteins after heat-shock.
Hul5 Ubiquitinates Proteins after Misfolding
We found that Hul5 is a major ubiquitin ligase that participates in the cytosolic heat-shock ubiquitination response. To identify the ubiquitin ligase that targets misfolded proteins after heat-shock, we screened over 80 yeast deletion mutant strains using a heat-shock ubiquitination response assay. Each of these mutant strains contains a single deletion of a known or putative E3 gene. Surprisingly, we found that none of the ubiquitin ligases with known PQC function contributed significantly to the increased ubiquitination detected after heat-shock. In contrast, cells that lack the ubiquitin ligase Hul5 display significantly reduced poly-ubiquitination after heat-shock. This suggests that Hul5 is a major ubiquitin ligase that participates in the cytosolic heat-shock ubiquitination response.Citation14 Hul5 is one of the five ligases containing a HECT (homologous to E6AP carboxyl terminal) domain in yeast; it interacts with proteasome and promotes proteasomal processivity by elongating ubiquitin chains on proteasome substrates.Citation18,Citation19
The ubiquitin ligase activity of Hul5 is required for its PQC function. Similar to HUL5 deletion, we found that mutation of the Hul5 ligase active site in the HECT domain leads to both an impaired ubiquitination response and reduced cell fitness after heat-shock, consistent with a function for Hul5 in stress response.Citation14 We showed that Hul5 co-immunoprecipitates with both Ubc4 and Ubc5, which are the two major cytosolic E2 conjugating enzymes that are required for the degradation of heat-induced misfolded proteins.Citation20,Citation21 Similarly, we found that the increased ubiquitination upon heat-shock was completely abolished in cells carrying deletions of both the UBC4 and UBC5 genes.
Hul5 targets heat-induced misfolded proteins for degradation and ubiquitinates both newly–synthesized and long-lived proteins upon heat-shock. We found that the absence of Hul5 strongly impairs the degradation of short-lived misfolded proteins using pulse radio labeling.Citation14 Since mild heat-stress seems to mainly affect short-lived proteins,Citation21 we next determined if Hul5 solely targets newly–synthesized misfolded proteins in a similar manner to Rkr1 ligase. In contrast, we found that deletion of HUL5 also affects the ubiquitination of long-lived proteins under heat-stress, which indicates that Hul5 targets misfolded proteins irrespective of time since translation. Therefore, Hul5 is unlikely to be associated with the translation apparatus.
Hul5 is also required for the ubiquitination of misfolded proteins that is caused by the inactivation of the major yeast Hsp70 cytosolic folding machinery. When the activities of all four SSA chaperones (Ssa1- 4) are inhibited, we observed an increased poly-ubiquitination that is mostly Hul5-dependent.Citation14 Interestingly, previous studies showed that the degradation of several misfolded proteins is SSA chaperone-dependent.Citation22-Citation24 For example, the degradation of the mammalian tumor-suppressor protein VHL (von Hippel-Lindau) in yeast requires the SSA chaperones and the Sti1 and Sse1 co-factors.Citation22 In this case, the SSA chaperones may promote the ubiquitination of misfolded proteins. Interestingly, the increased ubiquitination occurs in the absence of SSA activity in our experiments. Therefore, the Hul5 PQC pathway may not be dependent on the SSA chaperones, at least for a subset of targets. Collectively, we found that Hul5 is important for the ubiquitination and degradation of misfolded proteins and is part of a novel PQC pathway.
Hul5 Ubiquitinates Low-solubility Cytosolic Proteins
We reasoned that misfolded proteins were more likely to be insoluble and found that Hul5 specifically targets cytosolic proteins that are found in the low-solubility cellular fraction. Using quantitative mass spectrometry, we next sought to identify which proteins are ubiquitinated by Hul5. More specifically, we compared two cell populations to identify which proteins are less ubiquitinated in the absence of Hul5 (i.e., corresponding to its substrates).Citation14 We performed this experiment both after heat-shock and in unstressed cells, and we deduced the following three conclusions. First, the Hul5-candidate substrates we identified are mainly cytosolic proteins, which confirms that Hul5 plays a major role in a cytosolic PQC pathway. Second, Hul5-candidate substrates were also identified in the absence of heat-shock, which indicates that Hul5 function is also important in unstressed conditions. Lastly, the absence of Hul5 specifically affects the ubiquitination of low-solubility proteins but does not significantly affect the overall ubiquitination levels in the whole proteome (in both stressed and unstressed cells). We verified this observation by testing several candidate substrates of Hul5 such as the Pin3 prion-like protein. We found that after heat-shock, the majority of Pin3 becomes insoluble and that Pin3 Hul5-dependent ubiquitination is only detected in the insoluble fraction. Taken together, these data suggest that Hul5 has a housekeeping role to specifically ubiquitinate low solubility cytosolic misfolded proteins.
Cytosolic Localization of Hul5 is Required for its PQC Function
Hul5 needs to be localized in the cytoplasm to target misfolded proteins and maintain cell fitness. Since a large portion of Hul5 localizes in the nucleus,Citation25 a major question was why the majority of Hul5 substrates we identified are cytosolic. For instance, does the Hul5-pathway require the shuttling of misfolded proteins to the nucleus similar to the Ubr1-San1 pathway? We found instead that the Hul5 ligase itself relocates from the nucleus to the cytoplasm in response to heat-shock.Citation14 By adding a nuclear localization signal (NLS) sequence to Hul5, we restricted Hul5 to the nucleus (both in unstressed and stressed cells). Constraint of Hul5 localization in the nucleus leads to a reduced ubiquitination response, reduced cell fitness after heat-shock, and the stabilization of a Hul5 substrate, which are similar to the phenotypes observed in the HUL5 deletion strain. These results indicate that Hul5 cytosolic localization is required for its PQC function, and that the ubiquitination of Hul5 substrates occurs in the cytoplasm. One possibility is that Hul5 re-distribution may allow it to rapidly target a large number of cytosolic misfolded proteins after heat-shock. In contrast, maintaining low levels of Hul5 in the cytoplasm in unstressed cells may prevent the premature degradation of transiently misfolded proteins.
Recruiting Hul5 to Low Solubility Cytosolic Proteins
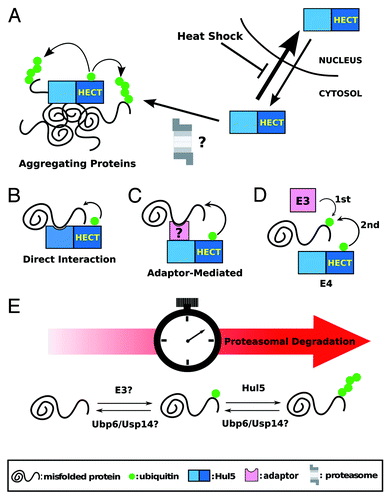
How is Hul5 recruited to the low-solubility cytosolic proteins? We found that Hul5 is enriched in the low solubility cellular fraction after heat-shock (N.N.F and T.M., unpublished data) suggesting that Hul5 may be directly recruited to protein aggregation sites in the cytoplasm (). Inhibition of the nuclear export machinery, using a Crm1/Xpo1 mutant, does not prevent the re-distribution of Hul5 in the cytoplasm (N.N.F, Vivien Measday and T.M., unpublished data). One possibility is that Hul5 constantly shuffles between the cytoplasm and the nucleus, and Hul5 nuclear import is blocked after heat-shock (). As well, Hul5 may be recruited together with the proteasome to the aggregation sites, since the proteasome has been shown to co-localize with the aggregatesCitation26 and Hul5 interacts with the proteasome.Citation18 Whether specific proteins promote Hul5 re-distribution and whether the PQC function of Hul5 depends on its interaction with the proteasome needs to be further studied. Since Hul5 promotes proteasome processivity,Citation19 its activity may also further assist substrate unfolding by increasing their affinity to the proteasome. Therefore, ubiquitination by Hul5 may facilitate the disaggregation of the low solubility proteins prior to their degradation.
Figure 1. (A) Schematic representation of Hul5 redistribution in the cell in response to heat-shock. (B-D) Schematic representations of the possible substrate recognition mechanisms by Hul5: Hul5 could directly recognize misfolded domains (B); an adaptor protein could tether the misfolded protein to Hul5 (C); and/or Hul5 could act as an E4 ligase by further ubiquitinating misfolded proteins first targeted by another E3 ligase (D). (E) A two-step ubiquitination of Hul5 substrates could act as a timer to only target proteins that are misfolded for an extended period of time for proteasomal degradation.
Recognition of Hul5 Substrates
A key question is how the Hul5-PQC recognizes misfolded proteins to be degraded. In addition to a possible Hul5 recruitment to protein aggregation sites, a specific recognition mechanism for Hul5 substrates must exist. One possibility is that Hul5 directly recognizes and binds to misfolded polypeptides (), similarly to the San1 E3 ligase that contains intrinsically-disordered regions that bind to misfolded domains.Citation27 Intriguingly, few nuclear proteins are targeted by Hul5 in unstressed cells, while Hul5 is mostly nuclear. Another factor, which specifically localizes in the cytoplasm, may be required to assist Hul5. This factor could either tether the misfolded substrate to Hul5 () or be another ubiquitin ligase (). Indeed, several Hul5 substrates that we verified remain mono-ubiquitinated in the absence of Hul5Citation14 suggesting that another E3 is involved in the process, and that Hul5 may work primarily as an ubiquitin chain assembly factor (also called E4), as previously suggested.Citation28 These three models are not mutually exclusive, and different combinations may be used for different substrates.
The Hul5-pathway may only target proteins that are misfolded or aggregated for an extended period of time. It was suggested that the deubiquitinating enzyme Ubp6 and Hul5 have antagonistic activities.Citation18 Interestingly, inhibition and mutation of the yeast Ubp6 and its mammalian Usp14 ortholog accelerate the degradation of aberrant proteins.Citation29,Citation30 Taken together, multiple steps in the ubiquitination of the low solubility proteins may act as a “timer” to ensure that only terminally misfolded proteins are targeted for degradation (). In this model, misfolded proteins are first ubiquitinated by another E3. Proteins that can refold or that are not permanently present in aggregation sites are de-ubiquitinated. In contrast, long-lasting low solubility proteins are eventually further ubiquitinated by Hul5 (due to its E4 activity) and targeted to the proteasome.
The Pin3 Case
Pin3/Lsb2 was identified as a putative Hul5 substrate by mass spectrometry.Citation14 We observed that heat-shock induces aggregation of Pin3, along with increased poly-ubiquitination that is Hul5-dependent. Pin3 is a prion-like protein that contains short stretches of glutamine residues and binds to actin patches. Overexpression of Pin3 promotes the conversion of the translation termination factor Sup35 into its prion form [PSI+]; while deletion of PIN3 destabilizes [PSI+].Citation31 Rsp5, another HECT domain-ubiquitin ligase, was shown to ubiquitinate Pin3 through a PY Rsp5-binding motif on Pin3. In this case, ubiquitination of Pin3 by Rsp5 reduces its ability to induce [PSI+] formation.Citation31 We confirmed that deletions of either HUL5 or RSP5 lead to the same phenotype after heat-shock, in which Pin3 poly-ubiquitination is abrogated (N.N.F and T.M., unpublished data). Therefore, Pin3 is likely first ubiquitinated by Rsp5 and then further ubiquitinated by the Hul5 E4 ligase. It will be interesting to determine whether the targeting of Pin3 by Hul5 requires Rsp5 itself, or only the ubiquitin attached to Pin3. Nevertheless, Rsp5 is most likely not the only ligase that adds the first ubiquitin moieties on all Hul5 substrates, since only a small portion of putative Hul5 substrates contain a PY motif.
Conclusion
The discovery of a Hul5-dependent PQC pathway confirms that multiple cellular strategies are in place to target cytosolic misfolded proteins. Hul5 is a 910-amino-acid-long protein with a C-terminal HECT domain. Around two-thirds of the Hul5 protein remains uncharacterized and future studies will have to focus on determining other functional domains. Moreover, it will be key to establish how Hul5 recognizes its PQC substrates.
Increased ubiquitination in mammalian cells upon heat-shock was observed over 25 y ago by the Rechteiner lab, but no enzymes have been characterized in this pathway. The human UBE3B and UBE3C ubiquitin ligases are two homologs that share the highest sequence similarity with Hul5. UBE3C (also called RAUL) is associated with nasal polyposisCitation32 and with the interferon response.Citation33 Future work is needed to investigate whether these two homologs of Hul5 also function in targeting misfolded proteins in mammalian cells.
| Abbreviations: | ||
| ERAD | = | endoplasmic-reticulum-associated protein degradation |
| NLS | = | nuclear localization signal |
| PQC | = | protein quality control |
Acknowledgments
The authors express gratitude to Dr. Measday (UBC) for comments on the manuscript and help with the Xpo1 experiment, Dr. Montpetit (UC, Berkeley) for the xpo1–1 strains, Adam Chruscicki (UBC) for comments on the manuscript, and other members of T.M. lab for their encouragement and discussion. We apologize for omitting colleague’s references due to the space limitation. T.M. is supported by a grant (MOP-89838) and a New Investigator Career Award from the Canada Institutes of Health Research (CIHR).
References
- McClellan AJ, Tam S, Kaganovich D, Frydman J. Protein quality control: chaperones culling corrupt conformations. Nat Cell Biol 2005; 7:736 - 41; http://dx.doi.org/10.1038/ncb0805-736; PMID: 16056264
- Wickner S, Maurizi MR, Gottesman S. Posttranslational quality control: folding, refolding, and degrading proteins. Science 1999; 286:1888 - 93; http://dx.doi.org/10.1126/science.286.5446.1888; PMID: 10583944
- Balch WE, Morimoto RI, Dillin A, Kelly JW. Adapting proteostasis for disease intervention. Science 2008; 319:916 - 9; http://dx.doi.org/10.1126/science.1141448; PMID: 18276881
- Vembar SS, Brodsky JL. One step at a time: endoplasmic reticulum-associated degradation. Nat Rev Mol Cell Biol 2008; 9:944 - 57; http://dx.doi.org/10.1038/nrm2546; PMID: 19002207
- Ravid T, Hochstrasser M. Diversity of degradation signals in the ubiquitin-proteasome system. Nat Rev Mol Cell Biol 2008; 9:679 - 90; http://dx.doi.org/10.1038/nrm2468; PMID: 18698327
- Rosenbaum JC, Gardner RG. How a disordered ubiquitin ligase maintains order in nuclear protein homeostasis. Nucleus 2011; 2; http://dx.doi.org/10.4161/nucl.2.4.16118; PMID: 21941105
- Connell P, Ballinger CA, Jiang J, Wu Y, Thompson LJ, Höhfeld J, et al. The co-chaperone CHIP regulates protein triage decisions mediated by heat-shock proteins. Nat Cell Biol 2001; 3:93 - 6; http://dx.doi.org/10.1038/35050618; PMID: 11146632
- Murata S, Minami Y, Minami M, Chiba T, Tanaka K. CHIP is a chaperone-dependent E3 ligase that ubiquitylates unfolded protein. EMBO Rep 2001; 2:1133 - 8; http://dx.doi.org/10.1093/embo-reports/kve246; PMID: 11743028
- Heck JW, Cheung SK, Hampton RY. Cytoplasmic protein quality control degradation mediated by parallel actions of the E3 ubiquitin ligases Ubr1 and San1. Proc Natl Acad Sci U S A 2010; 107:1106 - 11; http://dx.doi.org/10.1073/pnas.0910591107; PMID: 20080635
- Prasad R, Kawaguchi S, Ng DT. A nucleus-based quality control mechanism for cytosolic proteins. Mol Biol Cell 2010; 21:2117 - 27; http://dx.doi.org/10.1091/mbc.E10-02-0111; PMID: 20462951
- Sultana R, Theodoraki MA, Caplan AJ. UBR1 promotes protein kinase quality control and sensitizes cells to Hsp90 inhibition. Exp Cell Res 2012; 318:53 - 60; http://dx.doi.org/10.1016/j.yexcr.2011.09.010; PMID: 21983172
- Nillegoda NB, Theodoraki MA, Mandal AK, Mayo KJ, Ren HY, Sultana R, et al. Ubr1 and Ubr2 function in a quality control pathway for degradation of unfolded cytosolic proteins. Mol Biol Cell 2010; 21:2102 - 16; http://dx.doi.org/10.1091/mbc.E10-02-0098; PMID: 20462952
- Bengtson MH, Joazeiro CA. Role of a ribosome-associated E3 ubiquitin ligase in protein quality control. Nature 2010; 467:470 - 3; http://dx.doi.org/10.1038/nature09371; PMID: 20835226
- Fang NN, Ng AH, Measday V, Mayor T. Hul5 HECT ubiquitin ligase plays a major role in the ubiquitylation and turnover of cytosolic misfolded proteins. Nat Cell Biol 2011; 13:1344 - 52; http://dx.doi.org/10.1038/ncb2343; PMID: 21983566
- Derkatch IL, Bradley ME, Hong JY, Liebman SW. Prions affect the appearance of other prions: the story of [PIN(+)]. Cell 2001; 106:171 - 82; http://dx.doi.org/10.1016/S0092-8674(01)00427-5; PMID: 11511345
- Golebiowski F, Matic I, Tatham MH, Cole C, Yin Y, Nakamura A, et al. System-wide changes to SUMO modifications in response to heat shock. Sci Signal 2009; 2:ra24; http://dx.doi.org/10.1126/scisignal.2000282; PMID: 19471022
- Miller MJ, Barrett-Wilt GA, Hua Z, Vierstra RD. Proteomic analyses identify a diverse array of nuclear processes affected by small ubiquitin-like modifier conjugation in Arabidopsis. Proc Natl Acad Sci U S A 2010; 107:16512 - 7; http://dx.doi.org/10.1073/pnas.1004181107; PMID: 20813957
- Crosas B, Hanna J, Kirkpatrick DS, Zhang DP, Tone Y, Hathaway NA, et al. Ubiquitin chains are remodeled at the proteasome by opposing ubiquitin ligase and deubiquitinating activities. Cell 2006; 127:1401 - 13; http://dx.doi.org/10.1016/j.cell.2006.09.051; PMID: 17190603
- Aviram S, Kornitzer D. The ubiquitin ligase Hul5 promotes proteasomal processivity. Mol Cell Biol 2010; 30:985 - 94; http://dx.doi.org/10.1128/MCB.00909-09; PMID: 20008553
- Seufert W, Jentsch S. Ubiquitin-conjugating enzymes UBC4 and UBC5 mediate selective degradation of short-lived and abnormal proteins. EMBO J 1990; 9:543 - 50; PMID: 2154373
- Medicherla B, Goldberg AL. Heat shock and oxygen radicals stimulate ubiquitin-dependent degradation mainly of newly synthesized proteins. J Cell Biol 2008; 182:663 - 73; http://dx.doi.org/10.1083/jcb.200803022; PMID: 18725537
- McClellan AJ, Scott MD, Frydman J. Folding and quality control of the VHL tumor suppressor proceed through distinct chaperone pathways. Cell 2005; 121:739 - 48; http://dx.doi.org/10.1016/j.cell.2005.03.024; PMID: 15935760
- Park SH, Bolender N, Eisele F, Kostova Z, Takeuchi J, Coffino P, et al. The cytoplasmic Hsp70 chaperone machinery subjects misfolded and endoplasmic reticulum import-incompetent proteins to degradation via the ubiquitin-proteasome system. Mol Biol Cell 2007; 18:153 - 65; http://dx.doi.org/10.1091/mbc.E06-04-0338; PMID: 17065559
- Han S, Liu Y, Chang A. Cytoplasmic Hsp70 promotes ubiquitination for endoplasmic reticulum-associated degradation of a misfolded mutant of the yeast plasma membrane ATPase, PMA1. J Biol Chem 2007; 282:26140 - 9; http://dx.doi.org/10.1074/jbc.M701969200; PMID: 17631501
- Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, et al. Global analysis of protein localization in budding yeast. Nature 2003; 425:686 - 91; http://dx.doi.org/10.1038/nature02026; PMID: 14562095
- Kaganovich D, Kopito R, Frydman J. Misfolded proteins partition between two distinct quality control compartments. Nature 2008; 454:1088 - 95; http://dx.doi.org/10.1038/nature07195; PMID: 18756251
- Rosenbaum JC, Fredrickson EK, Oeser ML, Garrett-Engele CM, Locke MN, Richardson LA, et al. Disorder targets misorder in nuclear quality control degradation: a disordered ubiquitin ligase directly recognizes its misfolded substrates. Mol Cell 2011; 41:93 - 106; http://dx.doi.org/10.1016/j.molcel.2010.12.004; PMID: 21211726
- Koegl M, Hoppe T, Schlenker S, Ulrich HD, Mayer TU, Jentsch S. A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell 1999; 96:635 - 44; http://dx.doi.org/10.1016/S0092-8674(00)80574-7; PMID: 10089879
- Torres EM, Dephoure N, Panneerselvam A, Tucker CM, Whittaker CA, Gygi SP, et al. Identification of aneuploidy-tolerating mutations. Cell 2010; 143:71 - 83; http://dx.doi.org/10.1016/j.cell.2010.08.038; PMID: 20850176
- Lee BH, Lee MJ, Park S, Oh DC, Elsasser S, Chen PC, et al. Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature 2010; 467:179 - 84; http://dx.doi.org/10.1038/nature09299; PMID: 20829789
- Chernova TA, Romanyuk AV, Karpova TS, Shanks JR, Ali M, Moffatt N, et al. Prion induction by the short-lived, stress-induced protein Lsb2 is regulated by ubiquitination and association with the actin cytoskeleton. Mol Cell 2011; 43:242 - 52; http://dx.doi.org/10.1016/j.molcel.2011.07.001; PMID: 21777813
- Pasaje CF, Kim JH, Park BL, Park JS, Uh ST, Kim MK, et al. UBE3C genetic variations as potent markers of nasal polyps in Korean asthma patients. J Hum Genet 2011; 56:797 - 800; http://dx.doi.org/10.1038/jhg.2011.104; PMID: 21881582
- Yu Y, Hayward GS. The ubiquitin E3 ligase RAUL negatively regulates type i interferon through ubiquitination of the transcription factors IRF7 and IRF3. Immunity 2010; 33:863 - 77; http://dx.doi.org/10.1016/j.immuni.2010.11.027; PMID: 21167755