Abstract
We previously showed that over production of a fusion protein in which the prion domain of Saccharomyces cerevisiae [PSI+] is connected to glutathione S-transferase (GST-Sup35NM) causes a marked decrease in the colony forming ability of Escherichia coli strain BL21 after reaching stationary phase. Evidence indicated that the observed toxicity was attributable to intracellular formation of fibrous aggregates of GST-Sup35NM. In this report, we describe the isolation of plasmids that encode mutant forms of GST-Sup35NM which do not confer the toxicity to E. coli strain BL21. Each of the four spontaneous mutant-forms of GST-Sup35NM obtained revealed amino acid substitutions. One substitution was located in the N domain, and the others in the M domain. Congo red binding assay indicated that none of these mutant protein underwent conformational alteration in vitro. From these results, we conclude that the M domain, in collaboration with the N domain, plays an essential role in aggregation of Sup35NM. In addition, our data demonstrate the usefulness of the E. coli expression system in studying aggregate-forming proteins.
Introduction
Saccharomyces cerevisiae [PSI+] was first identified by its association with the termination-suppressor phenotype (Cox 1971).Citation1 Later, it was found that the eRF3 termination factor encoded by the SUP35 gene undergoes conformational alteration, though at very low incidence, and forms fibrous aggregates.Citation2–Citation6 Since this feature is analogous to mammalian prion, a pathogen that causes disorders of the central nervous system,Citation7,Citation8 [PSI+] is called yeast prion; [URE3]Citation9,Citation10 and [PIN+]Citation11,Citation12 are two other S. cerevisiae prion-like factors known by now. [PSI+] is not only an attractive research object in itself, but is also a useful experimental model system for elucidating conformational alteration of mammalian prion and aggregate-forming proteins.
Sup35 (viz, eRF3 and [PSI+]) consists of 685 amino acids and is divided into three functional domains: the C domain (C-terminal 432 amino acids) is responsible for the eRF3 function; the N domain (N-terminal 123 amino acids) manifests conformational alteration and aggregation; and the 130 amino acid region between the N and C domains, which is rich in polar amino acids and conventionally referred to as the M domain, is simply thought to acts as a linker of the N and C domains.Citation13,Citation14 With intention to elucidate the mechanism of conformational alteration and aggregation of Sup35, we constructed an E. coli plasmid suitable for production of a fusion protein in which glutathione S-transferase (GST: EC2.5.1.18)Citation15 is connected to the N-terminus of Sup35NM.Citation16 We found that E. coli strain BL21 (defective in OmpT and Lon proteases) producing this fusion protein grew normally to the stationary phase, and that their colony-forming ability in the stationary phase decreased much more quickly than that of control strain. It was also found that this toxicity was attributable to intracellular formation of fibrous aggregates of GST-Sup35NM. Later, it was shown that the deficiency in OmpT protease in E. coli strain BL21 was responsible for the observed toxicity.Citation17
In the course of our studies, we have observed rare events, at a frequency of about 5% of trials, in which colony forming ability in the stationary phase shows an atypical profile. Here, we describe this rare phenomenon, and present evidence showing that this rare event is attributable to spontaneous mutations that cause amino acid substitutions in Sup35NM. We then discuss the importance of our findings as they indicate usefulness of the present experimental system in the study of aggregate-forming proteins.
Results and Discussion
As previously reported, E. coli strain BL21 transformed with plasmid pGEX-SUP35NM (producing GST-Sup35NM) grew to the stationary phase following overnight culturing, after which the colony forming ability of the cells decreased rapidly (, solid line). However, we observed eight cases (approximately 5% of trials) in which colony forming ability during the stationary phase showed an atypical profile. That is, colony forming ability temporarily increased after 4–5 days of incubation (, dotted lines). For each of the eight exceptional cases, we picked a representative colony from a plate inoculated with a five day-old culture and inoculated into fresh LB medium containing Amp and IPTG. As shown in , the cells grew up normally to the stationary phase, and the colony forming ability of the cells was sustained at the same level for five days; the results of three representative clones are shown in .
It maybe worth mentioning that the colony forming ability of the primary cultures only temporary increased in the prolonged cultivation (see ), whereas that of the re-inoculated cultures sustained at higher level during the prolonged cultivation. The reason for this difference in behavior of the colony forming ability in the prolonged cultivation remains obscure. A possibility is that the primary cultures contain substance(s) that is excreted from the aggregate-containing cells and manifests inhibition of proliferation of the aggregate-free cells, but more investigation is certainly needed to elucidate this potentially interesting phenomenon.
Anyhow, we recovered plasmids from these eight clones (M-1 through M-8) and transformed E. coli strain BL21 with each. In all cases, the transformants grew to the stationary phase after overnight cultivation, and the colony forming ability of the cells was sustained at the same level for five days (). Hence, we concluded that the obtained eight mutants retained plasmids, but the plasmids no longer conferred the rapid decrease in colony forming ability to E. coli strain BL21. Next, we prepared cell extracts from these eight mutants and performed SDS-PAGE (). Four of the eight mutants (M-2, M-6, M-7 and M-8) revealed an intense band (indicated by solid arrow head) at a position corresponding to 55 kDa. BL21/pGEX-SUP35NM (lane C) also showed an intense band (indicated by open arrow head) at the same position, which was attributed to GST-Sup35NM.Citation16,Citation17 In contrast, the remaining four mutants (M-1, M-3, M-4 and M-5) did not exhibit the 55 kDa band, indicating that these mutants did not produce GST-Sup35NM. In contrary, M-2, M-6, M-7 and M-8 were concluded to produce mutant-forms of GST-Sup35NM.
We then carried out Congo red binding assay using the cell extract of M-2, M-6, M-7 and M-8 and found that they did not show increase in bound Congo red; the cell extract prepared from BL21/pGEX-SUP35NM showed a linear increase of bound Congo red as incubation time increased (). We next sequenced the SUP35NM region of the plasmids recovered from M-2, M-6, M-7 and M-8 (). While plasmid recovered from M-2, M-7 and M-8 contained single base substitutions of A143 with G, T431 with C and G720 with T, respectively, plasmid from M-6 comprised two independent base substitutions, T385 with with G and T440 with G. The presumed amino acid changes in the deduced amino acid sequence of Sup35NM caused by these base substitutions are illustrated in . M-2 encoded a single amino acid substitution, N48S, in the N domain, a position situated within the PAGGYYQNY sequence which appears as a six-time repeat and is thought to play a central role in stabilization of aggregate of Sup35 ().Citation22–Citation26
The remaining amino acid substitutions unveiled in this study (F129V, L144S, V147G and K240N) were located within the M domain (). Although the M domain is conventionally thought to be inert in conformational alteration and aggregation of Sup35 (see Introduction), more recent studies have indicated a vital function of the M domain in aggregation of Sup35.Citation27,Citation28 We have also reported that GST-Sup35NM cleaved by the OmpT protease at KK pairs in the M domain forms amorphous aggregates that are nontoxic to E. coli strain BL21.Citation17 The results presented in this report, together with these lines of information, indicate that M domain plays an essential role in aggregation of Sup35.
Sup35NM does not readily form crystals, and thus X-ray crystallography data are not available. Therefore, our argument on the impact of these amino acid substitutions on tertiary structure of the molecule is not practical. We thus limit our argument to the secondary structure of the molecule. shows features of the secondary structure of Sup35NM predicted by the method of Chou and Fasman.Citation29 M-6 comprised two amino acid substitutions, F129V and V147G. Although it remains to be determined whether either or both of these two amino acid substitutions are responsible for the loss of detrimental growth function, these substitutions do not cause charge alterations, nor are they affect Sup35NM secondary structure as predicted by the method of Chou and Fasman.Citation29 Therefore, we hypothesize that a very small distortion of the peptide backbone may affect aggregation of Sup35NM. In M-7, the single amino acid substitution L144S represents an alteration in charge. Interestingly, position 144 is located within the same predicted coil structure as position 147, leading to a speculation that M-6 mutation V147G is more likely to be responsible for the loss of function than F129V which is located within a predicted a-helix structure. M-8 encodes amino acid substitution K240N. Position 240 is located within a predicted long α-helix structure at the C-terminus of the M domain. According to the method of Chou and Fasman,Citation29 K240N is not predicted to affect the secondary structure of the region. However, since K240N confers a charge alteration, we contend that a slight stress in the peptide backbone is vital for aggregation of Sup35NM.
In the study of [PSI+], the words “mutant” and “strain” are usually used to refer to variant conformational forms of Sup35,Citation30–Citation32 because mutations due to base substitution of SUP35 (and thus amino acid substitution of Sup35) are rarely detected. In this report, we have shown that mutant forms of Sup35NM are relatively easily obtained in an experimental system in which Sup35NM is allowed to over-produce in E. coli strain BL21.Citation16,Citation17 With this experimental system, it is possible to accumulate many more mutant forms of Sup35NM, which will provide clues for understanding conformational alteration and aggregation of this molecule. Of additional significance, we suggest that this experimental system would be applicable to the study of many other aggregate-forming proteins.
Materials and Methods
Strains, growth media and growth conditions.
Escherichia coli strain BL21 (F-dcm ompT hsdSB(rB- mB-) gal)Citation18 was used exclusively for production of GST-Sup35NM in this study. Plasmid pGEX-SUP35NMCitation16 was used to direct production of GST-Sup35NM in E. coli. LB medium (1% bacto-tryptone, 0.5% yeast extract and 0.5% NaCl, pH 7.2) was used to grow E. coli. To ensure retention of plasmids, 50 µg/ml ampicillin (Amp) was added to LB medium. To stimulate the tac promoter, 24 µg/ml IPTG was added. The cells were cultured at 37°C in a rotary shaker at 120 rpm. For solid medium, 1% agar was added, and cells on the solid medium were grown at 37°C without shaking.
Measurements of colony-forming ability.
A volume of 20 ml culture medium in a 100 ml flask was inoculated with a 0.1 ml aliquot of pre-cultured cells (cells grown in LB medium for 5 to 10 h). Throughout the experimental period, 0.5 mg IPTG and 1 mg Amp were added daily to the culture. A 0.2 ml aliquot of the culture was spread onto agar LB medium; when necessary, an appropriate dilution was made prior to spreading. The plate was incubated overnight at 37°C, and the colonies were counted.
DNA manipulation.
Standard DNA manipulation proceduresCitation19 were used. For nucleotide sequencing, appropriate primers, 20mers containing nucleotide sequences within the SUP35NM gene, were used. DNA for sequence analysis was prepared using a DYEnamic Terminator Cycle Sequencing Kit (Amersham Pharmacia, Piscataway, NJ, USA) and analyzed using a DNA sequencer (ABI 373A; Applied Biosystems Japan Co., Ltd., Tokyo, Japan). Data analysis was performed using ABI 373A software (Applied Biosystems Japan Co., Ltd., Tokyo, Japan).
Preparation of cell extracts.
Cells were harvested by centrifugation, washed twice with sonication buffer (50 mM Tris-HCl pH 8.0, 50 mM NaCl and 1 mM EDTA), and suspended in an equal volume (w/v) of the same buffer. The cell suspension was sonicated with a Microson™ cell disruptor (Heat Systems Inc., Plainview, NY, USA); a cycle of 10 s sonication and 30 s pose was repeated 10 times. Cell homogenate was centrifuged at 25.7 × 103 g for 10 min to obtain the supernatant which was used as the cell extract.
Protein assay.
Protein concentrations in cell extracts were measured as described by Bradford,Citation20 using BSA as a control.
SDS-PAGE.
For SDS-PAGE, 10% acrylamide gel (10% acrylamide, 0.3% methylene bis-acrylamide, 0.4% Tris-HCl pH 8.8, 0.1% SDS, 0.04% APS, and 0.06% TEMED) was applied with an aliquot of the cell extract (100 µg protein). Electrophoresis was done under a constant current of 20 mA/gel (2 mm × 150 mm).
Congo red binding assay.
Cell extract was adjusted to 0.2 mg protein/ml with buffer C (5 mM potassium phosphate pH 7.4, and 150 mM NaCl) and incubated at 23°C with gentle shaking (2 strokes per min). Aliquots (0.3 ml) were removed at intervals, mixed with an equal volume of Congo red solution (20 µM Congo red, 5 mM potassium phosphate pH 7.4, and 150 mM NaCl), and measured for absorption at 540 nm (A540) and 477 nm (A477). Bound Congo red (CRB) was then estimated by the following equation: CRB = (A540/25,292) − (A477/46,306).Citation21
Prediction of peptide secondary structure.
For prediction of secondary structure of peptides, we adopted the methods of Chou and Fasman.Citation29 In practice, a program accessible via internet (http://mbs.cbrc.jp/papia-cgi/ssp_queryJ.pl.) was used.
Figures and Tables
Figure 1 Detection of mutants that did not exhibit rapid decrease of colony forming ability in the stationary phase. (A) E. coli strain BL21 was transformed with pGEX-SUP35NM, and colony forming ability was examined as described in Materials and Methods. In a majority of cases colony forming ability decreased quickly (solid line), but there were cases (at a frequency of about a 5% of the trials) in which the colony forming ability temporarily increased; of a total of eight such cases, three are shown in this figure (dotted lines). (B) From each of the eight exceptional cases, a representative clone was picked, inoculated in fresh growth medium, and examined for colony forming ability as before; only the three clones corresponding to the exceptional cases shown in are shown.
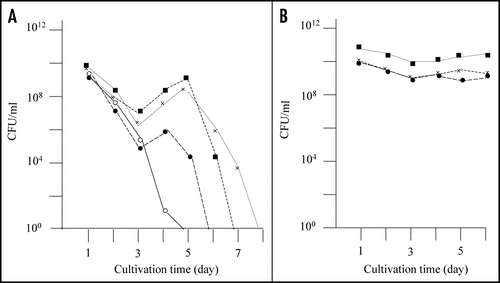
Figure 2 Nontoxic effects of the recovered plasmids against E. coli strain BL21. Colony forming ability of E. coli strain BL21 transformed with the recovered plasmids was examined; the results of three plasmids out of the eight recovered plasmids are included in this figure (solid marks). Colony forming ability of E. coli strain BL21 transformed with pGEX-SUP35NM is shown for comparison (open circle).
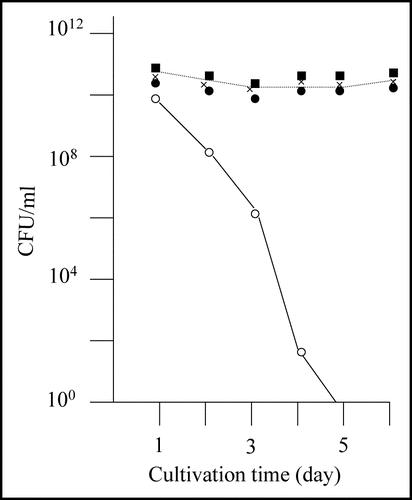
Figure 3 SDS-PAGE profiles of the eight spontaneous mutants obtained. Cell extract from each of the eight mutants of interest (M-1 through M-8) was prepared and subjected to SDS-PAGE as described in Materials and Methods. C and G represent control BL21/pGEX-SUP35NM (producing GST-Sup35NM) and BL21/pGEX-4T-3 (producing GST), respectively. SM1 and SM2 are two different mixtures of size markers. A band corresponding to a position of 55 kDa is marked with open or solid arrow head; for explanation, see text.
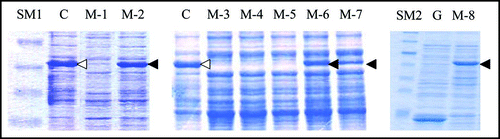
Figure 4 Congo red binding assay. The results of Congo red binding assay of the mutants, M-2 (▲),M-6 (●), M-7 (▽) and M-8 (■). The result of BL21/pGEX-SUP35NM (◯) is shown for comparison. For experimental details, see Materials and Methods.
Figure 5 Base substitutions detected in plasmid recovered from mutants. For each mutant, only the portion of relevant comparative SUP35NM sequence is shown. Numbers indicate base positions in SUP35NM. Letters below corresponding vertical arrows represent bases that were observed in the mutants.
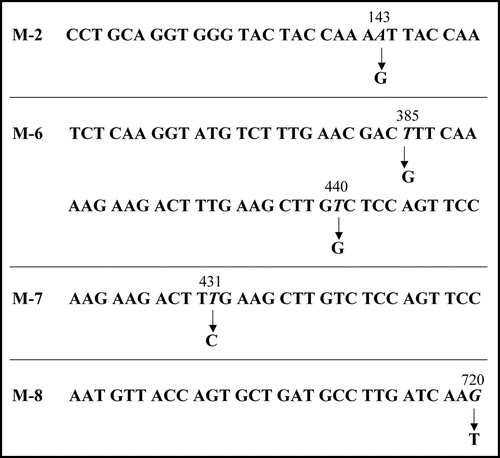
Figure 6 Amino acid replacement expected from detected base substitutions. Boxes represent the N and M domains of Sup35NM, and numbers below the boxes indicate amino acid positions in Sup35NM.
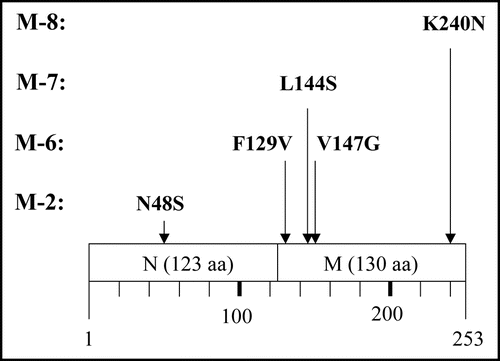
Figure 7 Predicted secondary structures of Sup35NM. Gothic and italic letters indicate amino acids in the N and M domains of Sup35, respectively; underscores indicate six times repeat of PQGGYQQYN present in the N domain of Sup35. Secondary structures within Sup35NM predicted by the method of Chou and Fasman (1978) are notated below the corresponding amino acid sequence; circle, square and triangle represent amino acids composing α helix, β Sheet and coil, respectively. Numbers indicate positions where amino acid replacements were determined in the present study.

Acknowledgements
This work was supported by funds from the Bio-Venture project and the 21st century COE program of Ritsumeikan University. HK was supported by the Frontier financial program of Ritsumeikan University.
References
- Cox BS. A recessive lethal super-suppressor mutation in yeast and other psi phenomena. Heredity 1971; 26:211 - 232
- Paushkin SV, Kushnirov VV, Smirnov VN, Ter-Avanesyan MD. Propagation of the yeast prion-like [PSI+] determinant is mediated by oligomerization of the SUP35-encoded polypeptide chain release factor. EMBO J 1996; 15:3127 - 3134
- Patino MM, Liu JJ, Glover JR, Lindquist S. Support for the prion hypothesis for inheritance of a phenotypic trait in yeast. Science 1996; 273:622 - 626
- Glover JR, Kowal AS, Schirmer EC, Patino MM, Liu JJ, Lindquist S. Self-seeded fibers formed by Sup35, the protein determinant of [PSI+], a heritable prion-like factor of S. cerevisiae. Cell 1997; 89:811 - 819
- Kushnirov VV, Ter-Avanesyan MD. Structure and replication of yeast prions. Cell 1998; 94:13 - 16
- Balbirnie M, Grothe R, Eisenberg DS. An amyloid-forming peptide from the yeast prion Sup35 reveals a dehydrated beta-sheet structure for amyloid. Proc Natl Acad Sci USA 2001; 98:2375 - 2380
- Prusiner SB. Prions are novel infectious pathogens causing scrapie and Creutzfeldt-Jakob disease. Bioessays 1986; 5:281 - 286
- Nelson R, Sawaya MR, Balbirnie M, Madsen AO, Riekel C, Grothe R, Eisenberg D. Structure of the cross-beta spine of amyloid-like fibrils. Nature (London) 2005; 435:773 - 777
- Derkatch IL, Bradley ME, Zhou P, Chernoff YO, Liebman SW. Genetic and environmental factors affecting the de novo appearance of the [PSI+] prion in Saccharomyces cerevisiae. Genetics 1997; 147:507 - 519
- Wickner RB. [URE3] as an altered URE2 protein: evidence for a prion analog in Saccharomyces cerevisiae. Science 1994; 264:566 - 569
- Masison DC, Wickner RB. Prion-inducing domain of yeast Ure2p and protease resistance of Ure2p in prion-containing cells. Science 1995; 270:93 - 95
- Derkatch IL, Bradley ME, Zhou P, Chernoff YO, Liebman SW. Genetic and environmental factors affecting the de novo appearance of the [PSI+] prion in Saccharomyces cerevisiae. Genetics 1997; 147:507 - 519
- Derkatch IL, Bradley ME, Masse SV, Zadorsky SP, Polozkov GV, Inge-Vechtomov SG, Liebman SW. Dependence and independence of [PSI(+)] and [PIN(+)]: a two-prion system in yeast?. EMBO J 2000; 19:1942 - 1952
- Kushnirov VV, Ter-Avanesyan MD, Telckov MV, Surguchov AP, Smirnov VN, Inge-Vechtomov SG. Nucleotide sequence of the SUP2 (SUP35) gene of Saccharomyces cerevisiae. Gene 1988; 66:45 - 54
- Ter-Avanesyan MD, Kushnirov VV, Dagkesamanskaya AR, Didichenko SA, Chernoff YO, Inge-Vechtomov SG, Smirnov VN. Deletion analysis of the SUP35 gene of the yeast Saccharomyces cerevisiae reveals two non-overlapping functional regions in the encoded protein. Mol Microbiol 1993; 7:683 - 692
- Habig WH, Pabst MJ, Jakoby WB. Glutathione S-transferases. The first enzymatic step in mercapturic acid formation. J Biol Chem 1974; 249:7130 - 7139
- Ono B, Kubota M, Kimiduka H, Kawaminami H, Ueto T, Yokosawa S, Iseda M, Yamamoto Y, Murakami Y, Yokota S. Production of a polymer forming fusion protein in Escerichia coli strain BL21. Biosci Biotechnol Biochem 2006; 70:60047 - 60051
- Ono B, Kimiduka H, Kubota M, Okuno K, Yabuta M. Role of the ompT mutation in stimulated decrease of colony forming ability due to intracellular protein aggregate formation in Escherichia coli strain BL21. Biosci Biotechnol Biochem 2007; 71:504 - 512
- Studier FW, Moffatt BA. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned gene. J Mol Biol 1986; 189:113 - 130
- Sambrook J, Fritsch EF, Maniatis T. Molecular Clonig: A Laboratory Manual 1989; 3rd ed Cold Spring Harbor Cold Spring Harbor Laboratory 31 - 34
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 1976; 72:248 - 254
- Klunk WE, Pettergrew JW, Abraham DJ. Quantitative evaluation of Congo red binding to amyloid-like proteins with a β-pleated sheet conformation. J Histchem Cytochem 1989; 37:1273 - 1281
- King CY, Tittmann P, Gross H, Gebert R, Aebi M, Wüthrich K. Prion-inducing domain 2–114 of yeast Sup35 protein transforms in vitro into amyloid-like filaments. Proc Natl Acad Sci USA 1997; 94:6618 - 6622
- Parham SN, Resende CG, Tuite MF. Oligopeptide repeats in the yeast protein Sup35p stabilize intermolecular prion interactions. EMBO J 2001; 20:2111 - 2119
- Chae YK, Cho KS, Chun W. A prionogenic peptide derived from Sup35 can force the whole GST fusion protein to show amyloid characteristics. Protein Pept Lett 2002; 9:315 - 321
- Resende CG, Outeiro TF, Sands L, Lindquist S, Tuite MF. Prion protein gene polymorphisms in Saccharomyces cerevisiae. Mol Microbiol 2003; 49:1005 - 1017
- Shkundina IS, Kushnirov VV, Tuite MF, Ter-Avanesyan MD. The role of the N-terminal oligopeptide repeats of the yeast Sup35 prion protein in propagation and transmission of prion variants. Genetics 2006; 172:827 - 835
- Liu JJ, Sondheimer N, Lindquist SL. Changes in the middle region of Sup35 profoundly alter the nature of epigenetic inheritance for the yeast prion [PSI+]. Proc Natl Acad Sci USA 2002; 99:16446 - 16453
- Bradley ME, Liebman SW. The Sup35 domains required for maintenance of weak, strong or undifferentiated yeast [PSI+] prions. Mol Microbiol 2004; 51:1649 - 1659
- Chou PY, Fasman GD. Empirical predictions of protein conformation. Ann Rev Biochem 1978; 47:251 - 276
- Zhou P, Derkatch IL, Uptain SM, Patino MM, Lindquist S, Liebman SW. The yeast non-Mendelian factor [ETA+] is a variant of [PSI+], a prion-like form of release factor eRF3. EMBO J 1999; 18:1182 - 1191
- Uptain SM, Sawicki GJ, Caughey B, Lindquist S. Strains of [PSI(+)] are distinguished by their efficiencies of prion-mediated conformational conversion. EMBO J 2001; 20:6236 - 6245
- Bradley ME, Edskes HK, Hong JY, Wickner RB, Liebman SW. Interactions among prions and prion “strains” in yeast. Proc Natl Acad Sci USA 2002; 99:16392 - 16399