Abstract
The study of fungal prion proteins affords remarkable opportunities to elucidate both intragenic and extragenic effectors of prion propagation. The yeast prion protein Sup35 and the self-perpetuating [PSI+] prion state is one of the best characterized fungal prions. While there is little sequence homology among known prion proteins, one region of striking similarity exists between Sup35p and the mammalian prion protein PrP. This region is comprised of roughly five octapeptide repeats of similar composition. The expansion of the repeat region in PrP is associated with inherited prion diseases. In order to learn more about the effects of PrP repeat expansions on the structural properties of a protein that undergoes a similar transition to a self-perpetuating aggregate, we generated chimeric Sup35-PrP proteins. Using both in vivo and in vitro systems we described the effect of repeat length on protein misfolding, aggregation, amyloid formation, and amyloid stability. We found that repeat expansions in the chimeric prion proteins increase the propensity to initiate prion propagation and enhance the formation of amyloid fibers without significantly altering fiber stability.
We recently described a novel chimeric prion system that was designed to elucidate the consequences of one class of inherited prion disease mutations on protein folding.Citation1,Citation2 We created a fusion between the mammalian prion protein PrP and the yeast prion protein Sup35p (). Sup35p is an essential translation termination factor in yeast. Interestingly, the majority of the protein can be sequestered into a self-propagating aggregate, the [PSI+] prion.Citation3 Remarkably, when yeast are grown in normal laboratory conditions, the [PSI+] prion is not detrimental. In fact, the biological consequences of the switch from the [psi−] non-prion state to the [PSI+] prion state may be beneficial in terms of adaptation and evolution.Citation4 Importantly, the prion state of Sup35p can be readily detected in vivo by monitoring the reduced function of the translation termination factor when the protein is propagating as a prion aggregate.Citation3 In addition, several methods have been developed to not only follow the propagation of the prion, but also to control the propagation and promote prion induction and loss (curing).Citation5 Therefore, in addition to simply being a fascinating biological problem in of itself, the [PSI+] prion in yeast affords the ability to further elucidate both intragenic and extragenic effectors of prion biology.
Several prions have now been identified and interestingly, there is little sequence homology between the proteins to suggest that only one type of sequence can form a self-propagating aggregate.Citation6–Citation8 In vitro studies suggest that many proteins can form amyloids under the appropriate conditions.Citation9 The fact that only a small percentage of proteins propagate as prions in vivo may be partly a consequence of physiological conditions being adequate to promote amyloid formation with those particular sequences. It is unclear what the precise distinction between prion and amyloid is at this time, but localization alone may preclude some amyloidogenic proteins from being “prion proteins” per se.Citation10
The sequence context that permits a protein to adopt a prion conformation in vivo is unclear. Several of the identified prion proteins have a domain that is enriched in glutamine and asparagine (Q/N) residues, but this is not true of all prion proteins.Citation7 Our recent study demonstrates that the Q/N character of the Sup35p prion-forming domain can be significantly reduced, yet still propagate as a prion.Citation1 This was also found recently in another prion protein chimera created and expressed in yeast.Citation6 These studies suggest that the lack of stable secondary structure may be one of the defining features of a prion-forming domain. One of the striking sequence similarities that does exist between two prion proteins occurs in an oligopeptide repeat region found in Sup35p and PrP.Citation11 Previous data clearly demonstrated that the Sup35p repeats are important for [PSI+] prion propagation.Citation12–Citation15 The deletion of a single repeat from the wild type SUP35 sequence results in the loss of normal [PSI+] prion propagation.Citation12 Moreover, the addition of two extra repeats of Sup35p sequence served to enhance the formation of the [PSI+] prion.Citation13 The expansion of the analogous repeat domain in the mammalian prion protein PrP is associated with an inherited form of prion disease.Citation16 Since the repeat regions of Sup35p and PrP are similar in size and character, we wanted to determine if the Sup35p oligopeptide repeat region could be substituted with that of PrP. Indeed, the PrP repeats in the context of Sup35p supported the propagation of the [PSI+] prion in yeast.Citation1,Citation17 Strikingly, we found phenotypic changes that occurred in a repeat length-dependent manner that suggested that the repeat expansions associated with disease result in an increase in the aggregation propensity but do not necessarily dictate only one type of aggregate structure.Citation1
More recently, we verified some of these results in vitro.Citation2 These data are in agreement with other studies on the effect of repeat expansions.Citation18,Citation19 Taking the analysis one step further, we demonstrated that the stability of the amyloid fibers formed with the repeat-expanded proteins did not differ significantly. A very interesting observation that we made was that the formation of amyloid fibers by the longest repeat-expanded chimera (SP14NM) followed drastically different kinetics compared to the chimera containing the wild type number of repeats (SP5NM).Citation2 In unseeded reactions, SP14NM did not show a lag phase during the course of fiber formation whereas SP5NM displayed a characteristic lag phase. Furthermore, the morphology of the amyloid fibers visualized by EM was different between SP14NM and SP5NM. SP14NM fibers were curvy and clumped but SP5NM fibers were long and straight. The correlation between the kinetics and the morphology of amyloid formation of SP14NM and SP5NM is reminiscent of fibers formed by β2-microglobulin (β2m) protein in different conditions.Citation20 At pH 3.6, β2m formed curvy, worm-like fibers with no apparent lag phase. In contrast, long, straight fibers were formed at pH 2.5 and had a distinct lag phase. Analysis of the β2m fibers formed at pH 3.6 using mass spectrometric techniques identified species ranging from monomer to 13-mer. This suggested that the fibers were formed by monomer addition. On the other hand, oligomers larger than tetramers were not formed during fiber formation at pH 2.5. Based on these data the authors propose that β2m forms fibers in a nucleation-independent manner at pH 3.6, but fiber formation at pH 2.5 follows a nucleation-dependent mechanism. We suggest that the mechanism underlying SP5NM and repeat-expanded SP14NM fiber formation is similar to β2m fibers formed at pH 2.5 and pH 3.6, respectively. It will be interesting to determine if disease-associated mutations in amyloidogenic proteins alter the pathway whereby amyloid formation occurs and how that process plays a role in pathogenesis.
In our in vivo study,Citation1 we highlighted a unique feature of the longest Sup35-PrP chimera that related to the ability of the protein to adopt multiple self-perpetuating prion conformations more readily than wild type Sup35p. We suggest that this may be an important aspect of prion biology as it relates to inherited disease. If the repeat-expanded proteins can adopt multiple conformations that aggregate, then that may contribute to the large amount of variation observed in pathology and disease progression in this class of inherited prion diseases.Citation21,Citation22
We also found that the spontaneous conversion of the repeat-expanded Sup35-PrP chimera into a prion state was significantly increased. However, this conversion required another aggregated protein in vivo, the [RNQ+] prion. In vitro, the prion-forming domain of the chimera showed a similar trend with the longer repeat lengths enhancing the ability of the protein to form amyloid fibers. The chimera with repeat expansions (8, 11 or 14 repeats) formed fibers very quickly as compared to that with the wild type number of repeats (5). While this correlates with the in vivo data in that both systems demonstrate an increased level of conversion with the repeat expansion, the systems are very different with respect to their requirement for a different “seed” to initiate the prion conversion. So, how does the [RNQ+] prion influence [PSI+]? At the moment, that isn't entirely clear. Susan Liebman and colleagues discovered another epigenetic factor in yeast, [PIN+], which was important for the de novo induction of [PSI+].Citation23–Citation25 Several years later, the [RNQ+] prionCitation26 was found to be that factor in the commonly used [PSI+] laboratory strains, but they also found that the overexpression of other proteins could reproduce the effect.Citation25 Hence, [RNQ+] can be [PIN+], and may be the primary epigenetic element that influences [PSI+] induction in yeast, but need not be in every case. Two models were proposed to explain the ability of [RNQ+] to influence the induction of [PSI+].Citation25,Citation27 One suggested that there is a direct templating effect where the aggregated state of the Rnq1 protein in the [RNQ+] prion serves as a seed for the direct physical association and aggregation of Sup35p and initiates [PSI+]. The second postulated that there is an inhibitor of aggregation in cells that is titrated out by the presence of another aggregated protein. Recent experimental evidence suggests that the templating model may explain at least part of the mechanism of action behind the [RNQ+] prion inducing the formation of [PSI+].Citation28,Citation29
Why is [RNQ+] required for the in vivo conversion of the repeatexpanded chimera that forms amyloid on its own very efficiently in vitro? Interestingly, we found that the [RNQ+] prion per se is not required. We overexpressed the Rnq1 protein from a constitutive high promoter (pGPD-RNQ1) and found that Rnq1p aggregated in the cells but did not induce the [RNQ+] prion. That is, the cells were still [rnq−] and did not genetically transmit the aggregated state of the protein. However, even these non-prion aggregates of Rnq1p served to enhance the induction of the chimeric prions. Therefore, either the [RNQ+] prion or an aggregate of Rnq1 protein is sufficient, which is in line with previous studies that demonstrated that some proteins that aggregate when overexpressed can also enhance the induction of [PSI+].Citation25 Also of note, recent data suggests that the requirement of [RNQ+] for the induction of Sup35p aggregation in vivo can be overcome by very long polyglutamine or glutamine/tyrosine stretches fused to the non-prion forming domain of Sup35p.Citation30 These fusions may alter protein-protein interactions or destabilize the non-prion structure of Sup35p in such a manner that the [RNQ+] prion seed is no longer required to form [PSI+] de novo. Indeed, the non-polymerizing state of some of the fusion proteins was shown to be very unstable.
So, what is the important difference between our in vitro and in vivo systems in the prion conversion? Obviously there are many candidates. First, the full length Sup35 protein may alter the conversion properties since a large part of the molecule is the structured C terminal domain. The C terminal domain may influence the initiation of prion propagation in vivo and that is not a factor in the in vitro system. Second, the influences of co-translational folding and potentially some initial unfolding of the prion-forming domain are not present since the in vitro system starts with denatured protein. Third, the environmental influences are clearly different. The molecular crowding effects and chaperones that are required for prion propagation in vivo are not required for the formation of amyloid in vitro. Finally, it is unclear if amyloid structures similar to those formed with the prion-forming domain in vitro actually exist in yeast. Certainly there is some correlation between the structures since aggregated Sup35 protein from [PSI+] cell lysates can seed amyloid formation in vitroCitation31,Citation32 and the fibers formed in vitro can be transformed into [psi−] cells and cause conversion to [PSI+].Citation33 Nevertheless, we find it interesting that the expansion of the repeat region can have a tremendous effect on amyloid formation in vitro yet still cannot overcome the requirement for [RNQ+] for conversion in vivo. The presence of co-aggregating or cross-seeding proteins may play a role in the sporadic appearance or progression of neurodegenerative diseases and the interconnected yeast prions [RNQ+] and [PSI+] may provide a model system for elucidating the mechanism underlying such effects.
Figures and Tables
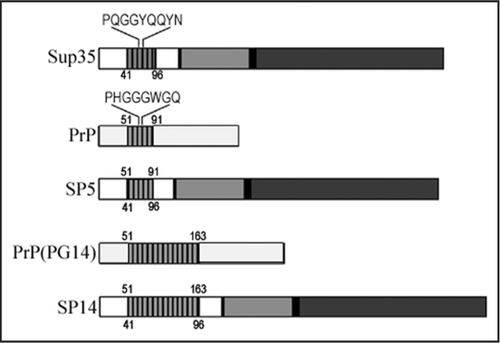
Figure 1 Schematic representation of the yeast protein Sup35p and the mammalian prion protein PrP highlighting the position of the oligopeptide repeat domain (ORD). The amino acid sequence represents the consensus for a single repeat. Numbers shown represent the amino acid position of the beginning and the end of each ORD. The numbers above the schematic represent the original PrP amino acid positioning and the numbers below represent the original Sup35p amino acid sequence positions.
References
- Tank EM, Harris DA, Desai AA, True HL. Prion protein repeat expansion results in increased aggregation and reveals phenotypic variability. Mol Cell Biol 2007; 27:5445 - 5455
- Kalastavadi T, True HL. Prion protein insertional mutations increase aggregation propensity but not fiber stability. BMC Biochem 2008; 9:7
- Tuite MF, Cox BS. Propagation of yeast prions. Nat Rev Mol Cell Biol 2003; 4:878 - 890
- True HL, Lindquist SL. A yeast prion provides a mechanism for genetic variation and phenotypic diversity. Nature 2000; 407:477 - 483
- Chernoff YO, Uptain SM, Lindquist SL. Analysis of prion factors in yeast. Methods Enzymol 2002; 351:499 - 538
- Taneja V, Maddelein ML, Talarek N, Saupe SJ, Liebman SW. A non-Q/N-rich prion domain of a foreign prion, [Het-s], can propagate as a prion in yeast. Mol Cell 2007; 27:67 - 77
- Uptain SM, Lindquist S. Prions as protein-based genetic elements. Annu Rev Microbiol 2002; 56:703 - 741
- Maddelein ML. Infectious Fold and Amyloid Propagation in Podospora anserina. Prion 2007; 1:44 - 47
- Dobson CM. The structural basis of protein folding and its links with human disease. Philos Trans R Soc Lond B Biol Sci 2001; 356:133 - 145
- Soto C, Estrada L, Castilla J. Amyloids prions and the inherent infectious nature of mis-folded protein aggregates. Trends Biochem Sci 2006; 31:150 - 155
- Bousset L, Melki R. Similar and divergent features in mammalian and yeast prions. Microbes Infect 2002; 4:461 - 469
- Parham SN, Resende CG, Tuite MF. Oligopeptide repeats in the yeast protein Sup35p stabilize intermolecular prion interactions. EMBO J 2001; 20:2111 - 2119
- Liu JJ, Lindquist S. Oligopeptide-repeat expansions modulate ‘protein-only’ inheritance in yeast. Nature 1999; 400:573 - 576
- Borchsenius AS, Wegrzyn RD, Newnam GP, Inge-Vechtomov SG, Chernoff YO. Yeast prion protein derivative defective in aggregate shearing and production of new ‘seeds’. EMBO J 2001; 20:6683 - 6691
- Osherovich LZ, Cox BS, Tuite MF, Weissman JS. Dissection and design of yeast prions. PLoS Biol 2004; 2:86
- Wadsworth JD, Hill AF, Beck JA, Collinge J. Molecular and clinical classification of human prion disease. Br Med Bull 2003; 66:241 - 254
- Dong J, Bloom JD, Goncharov V, Chattopadhyay M, Millhauser GL, Lynn DG, Scheibel T, Lindquist S. Probing the role of PrP repeats in conformational conversion and amyloid assembly of chimeric yeast prions. J Biol Chem 2007; 282:34204 - 34212
- Yu S, Yin S, Li C, Wong P, Chang B, Xiao F, et al. Aggregation of prion protein with insertion mutations is proportional to the number of inserts. Biochem J 2007; 403:343 - 351
- Moore RA, Herzog C, Errett J, Kocisko DA, Arnold KM, Hayes SF, Priola SA. Octapeptide repeat insertions increase the rate of protease-resistant prion protein formation. Protein Sci 2006; 15:609 - 619
- Smith AM, Jahn TR, Ashcroft AE, Radford SE. Direct observation of oligomeric species formed in the early stages of amyloid fibril formation using electrospray ionisation mass spectrometry. J Mol Biol 2006; 364:9 - 19
- Croes EA, Theuns J, Houwing-Duistermaat JJ, Dermaut B, Sleegers K, Roks G, et al. Octapeptide repeat insertions in the prion protein gene and early onset dementia. J Neurol Neurosurg Psychiatry 2004; 75:1166 - 1170
- King A, Doey L, Rossor M, Mead S, Collinge J, Lantos P. Phenotypic variability in the brains of a family with a prion disease characterized by a 144-base pair insertion in the prion protein gene. Neuropathol Appl Neurobiol 2003; 29:98 - 105
- Derkatch IL, Bradley ME, Masse SV, Zadorsky SP, Polozkov GV, Inge-Vechtomov SG, Liebman SW. Dependence and independence of [PSI(+)] and [PIN(+)]: a two-prion system in yeast. EMBO J 2000; 19:1942 - 1952
- Derkatch IL, Bradley ME, Zhou P, Chernoff YO, Liebman SW. Genetic and environmental factors affecting the de novo appearance of the [PSI+] prion in Saccharomyces cerevisiae. Genetics 1997; 147:507 - 519
- Derkatch IL, Bradley ME, Hong JY, Liebman SW. Prions affect the appearance of other prions: the story of [PIN(+)]. Cell 2001; 106:171 - 182
- Sondheimer N, Lindquist S. Rnq1: an epigenetic modifier of protein function in yeast. Mol Cell 2000; 5:163 - 172
- Osherovich LZ, Weissman JS. Multiple Gln/Asn-rich prion domains confer susceptibility to induction of the yeast [PSI(+)] prion. Cell 2001; 106:183 - 194
- Vitrenko YA, Gracheva EO, Richmond JE, Liebman SW. Visualization of aggregation of the Rnq1 prion domain and cross-seeding interactions with Sup35NM. J Biol Chem 2007; 282:1779 - 1787
- Derkatch IL, Uptain SM, Outeiro TF, Krishnan R, Lindquist SL, Liebman SW. Effects of Q/N-rich, polyQ, and non-polyQ amyloids on the de novo formation of the [PSI+] prion in yeast and aggregation of Sup35 in vitro. Proc Natl Acad Sci USA 2004; 101:12934 - 12939
- Alexandrov IM, Vishnevskaya AB, Ter-Avanesyan MD, Kushnirov VV. Appearance and propagation of polyglutamine-based amyloids in yeast: Tyrosine residues enable polymer fragmentation. J Biol Chem 2008; 283:15185 - 15192
- Paushkin SV, Kushnirov VV, Smirnov VN, Ter-Avanesyan MD. In vitro propagation of the prion-like state of yeast Sup35 protein. Science 1997; 277:381 - 383
- Glover JR, Kowal AS, Schirmer EC, Patino MM, Liu JJ, Lindquist S. Self-seeded fibers formed by Sup35, the protein determinant of [PSI+], a heritable prion-like factor of S. cerevisiae. Cell 1997; 89:811 - 819
- Tanaka M, Chien P, Naber N, Cooke R, Weissman JS. Conformational variations in an infectious protein determine prion strain differences. Nature 2004; 428:323 - 328