Abstract
Prions consist of PrPSc, a misfolded version of the cellular protein PrPC. They occur in a variety of strains that share the amino acid sequence of PrP but differ in phenotypic properties, such as cell tropism and pathogenicity; strain-ness is attributed to the conformation of PrPSc. To gain insight as to how susceptibility of cells to a given prion strain comes about, we compared amplification of RML prions by PMCA, using cell lysates from related, RML-resistant and RML-susceptible cell lines as substrate. We found that both lysates supported amplification of RML PrPSc equally well, despite a 280-fold difference in the susceptibility of the cells from which they were derived. Thus, susceptibility is an attribute of the intact cell.
Keywords: :
Introduction
Prion diseases are fatal neurodegenerative disorders affecting humans and many other mammals.Citation1 Well-known prion diseases include kuru and Creutzfeldt-Jakob disease in humans, scrapie in sheep, bovine spongiform encephalopathy (BSE or mad cow disease) in cattle and chronic wasting disease (CWD) in deer.
The hallmark of all TSEs is the conversion of the cellular protein, PrPC into a distinct isoform, PrPSc, which accumulates mainly in the brain and is the major, if not the only component of the prion. The proposal that PrPSc is the transmissible agent causing prionopathies and does not contain informational nucleic acids is supported by the cell-free synthesis of infectious prions from purified components, namely E.coli-derived, recombinant PrP in the presence of a phospholipid and poly(rA),Citation2 using Protein Misfolding Cyclic Amplification (PMCA).Citation3-Citation5
Interestingly, prions occur in the form of distinct strains which share the sequence of the underlying PrP but exhibit diverse phenotypes, including incubation period, clinical symptoms as well as biochemical and neuropathological features.Citation6 It is generally accepted that the differences between prion strains are based on distinctive conformations of the misfolded PrPSc, whose epigenetic characteristics are propagated with high fidelity (reviewed in refs. Citation7 and Citation8).
Strains were originally distinguished by their incubation periods and neuropathological characteristics in panels of inbred miceCitation9-Citation12in assays that required years to complete. The finding that cell lines showed differential susceptibilities to murine prion strainsCitation13-Citation16led to the development of the Cell Panel Assay (CPA),Citation17 which distinguishes many murine prion strains reliably and within less than 2 weeks by means of the Standard Scrapie Cell Assay (SSCA).Citation14
In this work we wanted to determine whether two closely related cell lines, R332H11 and PK1, which are both derived from murine N2a neuroblastoma cellsCitation14 but have very distinct susceptibilities to RML prions, differed in regard to their content of particular factors, or whether differential susceptibility was caused by processes dependent on the integrity of the cell, such as selective prion uptake or clearance. It has been shown previously that lysates of cultured cells can be used as a source of PrPC to amplify hamster- and mouse-adapted prion strains by Protein Misfolding Cyclic Amplification (PMCA).Citation18,Citation19 We therefore established PMCA using lysates of the RML-resistant R332H11Citation20 and RML-susceptible PK1 cells,Citation14 and showed that both lysates were equally efficient at amplifying RML prions. Therefore, the differential susceptibility of the two cell lines is dependent on the integrity of the cells, and PMCA is not a reliable predictor of cellular susceptibility to prion strains.
Results
The susceptibility of R332H11 and PK1 cells to 22L and RML prions was determined by the SSCA, as shown in . In short, cells in 96-well plates were exposed to serial dilutions of prion-infected brain homogenate, grown to confluence, and split three times. After reaching confluence, 20,000 cells from each well were transferred into Elispot plates and assayed for PrPSc-positive cells by ELISA.Citation21 The number of PrPSc-positive cells (“spots”) per 20,000 cells was plotted against the logarithm of the brain homogenate dilution and the RI1000 (Response Index1000, the reciprocal of the sample dilution giving 1000 spots) was determined. The ratio of RIs for RML on PK1 and R332H11 cells was 8.7 × 105/ 3.1 × 103 ≈280, confirming the relatively low susceptibility of R332H11 cells to infection by RML prions, while the RI ratio for 22L was 5.8.
Figure 1. PMCA is not predictive of cellular susceptibility to prion infection. Susceptibility of PK1 and R332H11 cells to RML and 22L prions was determined by the SSCA. PK1 and R332H11 cells were infected with serial 1:3 dilutions of (A) RML- or (B) 22L-infected brain homogenate and split 3 times. The number of PrPSc-positive cells per 20000 cells was determined by ELISA and plotted against the logarithm of the inoculum concentration. The RI1000 values (the reciprocals of the concentrations required to generate 1000 positive cells) are given in the upper left. Western blot showing that lysates of PK1 cells (19.6 mg protein/ml) and R332H11 cells (18.8 mg protein/ml) contained similar PrPC levels (18.6 and 17.7 arbitrary units, respectively) (C). (D) PMCA with cell lysates as source of PrPC and RML- or 22L-infected brain homogenates as seed. Triplicate samples from a representative experiment were displayed by western blotting (“-“, frozen controls; “+,” PMCA-amplified samples) (D). Average amplification, calculated from the ratio of intensities of PMCA-treated to frozen samples from four independent experiments for RML seed (left panel), was 4.9 ± 1.5 for PK1 and 5 ± 2.1 for R332H11 lysate. The average amplification for 22L seed (right panel), was 1.6 ± 0.8 for PK1 and 1.5 ± 0.3 for R332H11 lysate (E). Thus there was no significant difference between PMCA using PK1 or R332H11 lysates for either seed, as assessed by the Student’s t-test.
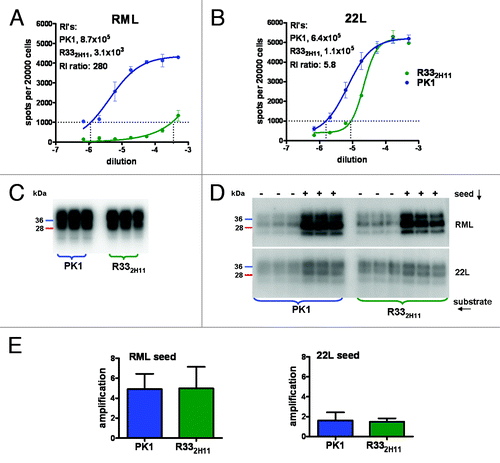
A prerequisite for susceptibility of a cell to infection by a prion strain is the presence of PrPC monomers with conformations permitting efficient accretion to the incoming PrPSc.Citation22–Citation25 To determine whether the PrPC of R332H11 cells was less efficiently converted to RML-specific PrPSc than that of PK1 cells, we prepared lysates from both cell lines and used them as substrate for RML PrPSc-directed PMCA. The two lysates had about equal levels of PrPC () and, surprisingly, were equally efficient in sustaining RML PrPSc synthesis (), leading on average to 5-fold amplification after 24 cycles of sonication/incubation (), a result that was obtained in four separate experiments. The 22L strain was poorly amplified with either lysate () showing an average of 1.5 fold amplification in four experiments (). Therefore, the differential susceptibility of R332H11 and PK1 cells to RML and 22L prions was not reflected in the PMCA based on their lysates.
Discussion
Transmission of prions between animal species expressing PrP genes with distinct sequences is in most cases inefficient. This so-called “species barrier” can frequently be abrogated by introducing donor PrP transgenes into the recipient, but in some cases this is not sufficient, implying that other factors are required.Citation26 Distinct cell lines show very different behaviors in regard to prion susceptibility. Rabbit-derived RK13 epithelial cells, which express very low levels of endogenous PrP, can be rendered susceptible to prions of other species by transfection with the cognate PrP genes, showing that in these cells PrPC is the critical exogenous factor.Citation27,Citation28 On the other hand different cell lines expressing the same PrP gene may exhibit opposite responsiveness to a particular prion strain,Citation17 and a given cell line may be able to discriminate between two different prion strains originating from the same species,Citation13,Citation15,Citation17,Citation29,Citation30 showing that factors other than the PrP sequence play a critical role.
Why can PK1 but not R332H11 cells replicate RML prions, although their lysates sustain PMCA-mediated replication of RML PrPSc with equal efficiency? Doubtless the mechanism operative in PMCA, believed to be breakage of PrPSc aggregates by sonication followed by accretion of PrPC to the newly generated surfaces, is very different from that in cells, where PrPC conversion, at least in yeast, is mediated by chaperones.Citation31 An additional reason could be that susceptibility is determined by processes linked to cellular integrity. Discrimination between strains at the level of prion uptake seems unlikely in view of the unspecific incorporation of PrPSc reported earlier,Citation32 however the relative rates of synthesis and degradation of PrPSc, which are critical for susceptibility to prion infection, may well depend on the integrity of the cell. In conclusion, PMCA, while being an immensely useful tool, is not predictive of cellular response to prions.
Materials and Methods
Cell culture
The isolation of cell lines has been described for PK1Citation14 and R332H11.Citation17 Cells were maintained in OBGS (Opti-Mem [Invitrogen], 4.5% BGS [Hyclone], 1% penicillin-streptomycin [Invitrogen]) and split 1:10 or 1:8 until they reached 80% confluence. After 7–8 splits they were discarded and a new batch of cells was thawed.
Prions
The RML isolate (RML I856-II) was obtained from the Prion Unit, University College London and was first propagated in CD1 and then in C57BL/6 mice (Charles River Laboratories). The 22L strain was obtained from the TSE Resource Centre, Compton, Newbury, UK and was propagated initially in CD1 and then in C57BL/6 mice. The titers, determined by end-point titrations were, in LD50 units/g brain, 108.75 for RML and 108.26 for 22L.Citation17
Brain homogenates
Frozen brains from mice infected with RML and 22L were homogenized in PBS (9 ml per g) using a ribolyser (FastPrep FP120, Bio 101, Thermo Electron Corp., Thermo Fischer Scientific) with ZrO beads (Glen Mills Inc.) at the 6.5 setting for 15 sec in Fast Prep tubes (MP Biomedicals). Homogenates were stored in small aliquots at −80°C. Thawed homogenates were syringed with a 28-gauge needle before use.
Standard Scrapie Cell Assay (SSCA)
PK1 and R332H11 cells were seeded at 5000 cells/well into 96-well plates and infected in triplicate with 1:3 serial dilutions of 5 × 10−4 diluted RML- and 22L-infected brain homogenates. After 4 d the cells were split 1:8, allowed to reach confluence and split 1:8 twice. After reaching confluence following the third split, 20000 cells from each well were transferred into Elispot plates (Millipore). PrPSc-positive cells were determined as previously described.Citation21 Briefly, the dried-down cells were lysed, digested with proteinase K, denatured with guanidinium chloride and subjected to an ELISA using the anti-PrP monoclonal antibody D18.Citation33 The number of spots per 20000 cells was determined with the Bioreader 5000 (BioSys), plotted against the logarithm of the homogenate dilution using GraphPad Prism and the RI1000 (reciprocal of the sample dilution yielding 1000 PrPSc-positive cells) was determined from the graph.
Protein Misfolding Cyclic Amplification (PMCA) using cell lysates as substrate
PK1 and R332H11 cells were grown in OBGS in 15-cm plates to 80% confluence, washed with cold PBS, collected in 10 ml PBS per plate on ice, counted and centrifuged at 4°C for 5 min at 3000 rpm. The pellet was suspended in Cell Conversion Buffer (150 mM NaCl, 1% Triton X-100, 5 mM EDTA in PBS with Protease Inhibitor Cocktail [Roche]) to give 4 × 107 cells/ml. Cells were lysed by syringing through a 28-gauge needle and centrifuged for 5 min at 4°C, 2000 g. The supernatants were collected and either frozen at -80°C until used or used immediately. Total protein content in the lysates was assayed by BCA (Pierce).
For the PMCA reactions 4 µl of 1% w/v RML infected brain homogenate in PBS was added as seed to 396 µl cell lysate (10−4 final dilution of infected brain homogenate). Six aliquots (60 µl) of each sample were distributed into PCR tubes (Axygen) containing 37 ± 3 mg of 1.0 mm Zirconia/Silica beads (Biospec products); three aliquots were frozen and three were subjected to cycles of 20 sec sonication and 30 min incubation at 37°C, using a Mixonic 3000 sonicator at the 8.5 power setting. After 12 h of PMCA, 20-µl aliquots of each sample were digested by adding 5 µl of 0.2 µg proteinase K/μl (Roche; 40 µg/ml final concentration) for 1 h with rotary shaking at 850 rpm and 56°C. The reaction was stopped by heating to 100°C with 6.5 µl of 4 × loading buffer (BioRad) for 10 min. Aliquots (15 µl) were electrophoresed through 4–12% polyacrylamide precast Criterion gels (BioRad) and PrP was displayed using 1 µg anti-PrP monoclonal D18/mlCitation33 and HPR anti-human secondary antibody (1:10000; Southern Biotech) followed by exposure to ECL plus (Pierce). Chemiluminiscence was recorded by CCD imaging (BioSpectrum AC Imaging System; UVP) and densitometric data were analyzed using Microsoft Excel and GraphPad Prism. PageRuler Plus Prestained Protein Ladder (Fermentas) was run as molecular weight marker.
Disclosure of Potential Conflicts of Interest
The authors declare that there no conflicts of interest.
Acknowledgments
We thank Anja M.Oelschlegel for help with the SSCA assays and for critical reading of the manuscript. This work was supported by grants from the National Institutes of Health 1RO1NSO59543, 1RO1NS067214 and from the Alafi Family Foundation to C.W.
Author contributions: M.E.H. performed all experiments, C.W. and M.E.H. conceived the experiments and wrote the manuscript.
References
- Prusiner SB. Molecular biology of prion diseases. Science 1991; 252:1515 - 22; http://dx.doi.org/10.1126/science.1675487; PMID: 1675487
- Wang F, Wang X, Yuan C-G, Ma J. Generating a prion with bacterially expressed recombinant prion protein. Science 2010; 327:1132 - 5; http://dx.doi.org/10.1126/science.1183748; PMID: 20110469
- Castilla J, Morales R, Saá P, Barria M, Gambetti P, Soto C. Cell-free propagation of prion strains. EMBO J 2008; 27:2557 - 66; http://dx.doi.org/10.1038/emboj.2008.181; PMID: 18800058
- Saá P, Castilla J, Soto C. Cyclic amplification of protein misfolding and aggregation. Methods Mol Biol 2005; 299:53 - 65; PMID: 15980595
- Saborio GP, Permanne B, Soto C. Sensitive detection of pathological prion protein by cyclic amplification of protein misfolding. Nature 2001; 411:810 - 3; http://dx.doi.org/10.1038/35081095; PMID: 11459061
- Bruce ME. TSE strain variation. Br Med Bull 2003; 66:99 - 108; http://dx.doi.org/10.1093/bmb/66.1.99; PMID: 14522852
- Morales R, Abid K, Soto C. The prion strain phenomenon: molecular basis and unprecedented features. Biochim Biophys Acta 2007; 1772:681 - 91; PMID: 17254754
- Weissmann C, Li J, Mahal SP, Browning S. Prions on the move. EMBO Rep 2011; 12:1109 - 17; http://dx.doi.org/10.1038/embor.2011.192; PMID: 21997298
- Bruce ME, McBride PA, Farquhar CF. Precise targeting of the pathology of the sialoglycoprotein, PrP, and vacuolar degeneration in mouse scrapie. Neurosci Lett 1989; 102:1 - 6; http://dx.doi.org/10.1016/0304-3940(89)90298-X; PMID: 2550852
- Fraser H, Bruce M. Argyrophilic plaques in mice inoculated with scrapie from particular sources. Lancet 1973; 1:617 - 8; http://dx.doi.org/10.1016/S0140-6736(73)90775-7; PMID: 4120692
- Fraser H, Dickinson AG. The sequential development of the brain lesion of scrapie in three strains of mice. J Comp Pathol 1968; 78:301 - 11; http://dx.doi.org/10.1016/0021-9975(68)90006-6; PMID: 4970192
- Dickinson AG, Meikle VM, Fraser H. Identification of a gene which controls the incubation period of some strains of scrapie agent in mice. J Comp Pathol 1968; 78:293 - 9; http://dx.doi.org/10.1016/0021-9975(68)90005-4; PMID: 4970191
- Bosque PJ, Prusiner SB. Cultured cell sublines highly susceptible to prion infection. J Virol 2000; 74:4377 - 86; http://dx.doi.org/10.1128/JVI.74.9.4377-4386.2000; PMID: 10756052
- Klöhn PC, Stoltze L, Flechsig E, Enari M, Weissmann C. A quantitative, highly sensitive cell-based infectivity assay for mouse scrapie prions. Proc Natl Acad Sci U S A 2003; 100:11666 - 71; http://dx.doi.org/10.1073/pnas.1834432100; PMID: 14504404
- Nishida N, Harris DA, Vilette D, Laude H, Frobert Y, Grassi J, et al. Successful transmission of three mouse-adapted scrapie strains to murine neuroblastoma cell lines overexpressing wild-type mouse prion protein. J Virol 2000; 74:320 - 5; http://dx.doi.org/10.1128/JVI.74.1.320-325.2000; PMID: 10590120
- Vorberg I, Raines A, Story B, Priola SA. Susceptibility of common fibroblast cell lines to transmissible spongiform encephalopathy agents. J Infect Dis 2004; 189:431 - 9; http://dx.doi.org/10.1086/381166; PMID: 14745700
- Mahal SP, Baker CA, Demczyk CA, Smith EW, Julius C, Weissmann C. Prion strain discrimination in cell culture: the cell panel assay. Proc Natl Acad Sci U S A 2007; 104:20908 - 13; http://dx.doi.org/10.1073/pnas.0710054104; PMID: 18077360
- Browning S, Baker CA, Smith E, Mahal SP, Herva ME, Demczyk CA, et al. Abrogation of complex glycosylation by swainsonine results in strain- and cell-specific inhibition of prion replication. J Biol Chem 2011; 286:40962 - 73; http://dx.doi.org/10.1074/jbc.M111.283978; PMID: 21930694
- Mays CE, Yeom J, Kang HE, Bian J, Khaychuk V, Kim Y, et al. In vitro amplification of misfolded prion protein using lysate of cultured cells. PLoS One 2011; 6:e18047; http://dx.doi.org/10.1371/journal.pone.0018047; PMID: 21464935
- Mahal SP, Browning S, Li J, Suponitsky-Kroyter I, Weissmann C. Transfer of a prion strain to different hosts leads to emergence of strain variants. Proc Natl Acad Sci U S A 2010; 107:22653 - 8; http://dx.doi.org/10.1073/pnas.1013014108; PMID: 21156827
- Mahal SP, Demczyk CA, Smith EW Jr., Klohn PC, Weissmann C. Assaying prions in cell culture: the standard scrapie cell assay (SSCA) and the scrapie cell assay in end point format (SCEPA). Methods Mol Biol 2008; 459:49 - 68; http://dx.doi.org/10.1007/978-1-59745-234-2_4; PMID: 18576147
- Collinge J, Clarke AR. A general model of prion strains and their pathogenicity. Science 2007; 318:930 - 6; http://dx.doi.org/10.1126/science.1138718; PMID: 17991853
- DeArmond SJ, Sánchez H, Yehiely F, Qiu Y, Ninchak-Casey A, Daggett V, et al. Selective neuronal targeting in prion disease. Neuron 1997; 19:1337 - 48; http://dx.doi.org/10.1016/S0896-6273(00)80424-9; PMID: 9427256
- Weissmann C. The state of the prion. Nat Rev Microbiol 2004; 2:861 - 71; http://dx.doi.org/10.1038/nrmicro1025; PMID: 15494743
- Weissmann C. Birth of a prion: spontaneous generation revisited. Cell 2005; 122:165 - 8; http://dx.doi.org/10.1016/j.cell.2005.07.001; PMID: 16051142
- Telling GC, Scott M, Mastrianni J, Gabizon R, Torchia M, Cohen FE, et al. Prion propagation in mice expressing human and chimeric PrP transgenes implicates the interaction of cellular PrP with another protein. Cell 1995; 83:79 - 90; http://dx.doi.org/10.1016/0092-8674(95)90236-8; PMID: 7553876
- Courageot MP, Daude N, Nonno R, Paquet S, Di Bari MA, Le Dur A, et al. A cell line infectible by prion strains from different species. J Gen Virol 2008; 89:341 - 7; http://dx.doi.org/10.1099/vir.0.83344-0; PMID: 18089759
- Vilette D, Andreoletti O, Archer F, Madelaine MF, Vilotte JL, Lehmann S, et al. Ex vivo propagation of infectious sheep scrapie agent in heterologous epithelial cells expressing ovine prion protein. Proc Natl Acad Sci U S A 2001; 98:4055 - 9; http://dx.doi.org/10.1073/pnas.061337998; PMID: 11259656
- Herva ME, Relaño-Ginés A, Villa A, Torres JM. Prion infection of differentiated neurospheres. J Neurosci Methods 2010; 188:270 - 5; http://dx.doi.org/10.1016/j.jneumeth.2010.02.022; PMID: 20206206
- Rubenstein R, Deng H, Race RE, Ju W, Scalici CL, Papini MC, et al. Demonstration of scrapie strain diversity in infected PC12 cells. J Gen Virol 1992; 73:3027 - 31; http://dx.doi.org/10.1099/0022-1317-73-11-3027; PMID: 1359002
- Doyle SM, Wickner S. Hsp104 and ClpB: protein disaggregating machines. Trends Biochem Sci 2009; 34:40 - 8; http://dx.doi.org/10.1016/j.tibs.2008.09.010; PMID: 19008106
- Magalhães AC, Baron GS, Lee KS, Steele-Mortimer O, Dorward D, Prado MA, et al. Uptake and neuritic transport of scrapie prion protein coincident with infection of neuronal cells. J Neurosci 2005; 25:5207 - 16; http://dx.doi.org/10.1523/JNEUROSCI.0653-05.2005; PMID: 15917460
- Williamson RA, Peretz D, Pinilla C, Ball H, Bastidas RB, Rozenshteyn R, et al. Mapping the prion protein using recombinant antibodies. J Virol 1998; 72:9413 - 8; PMID: 9765500