Abstract
Evidence is now accumulating that damaged proteins are not randomly distributed but often concentrated in microscopically visible and functionally distinct inclusion bodies. How misfolded proteins are organized into these compartments, however, is still unknown. We have recently begun to investigate stress-inducible protein quality control (PQC) bodies in yeast cells. Surprisingly, we found that protein misfolding and aggregation were not sufficient to trigger body formation under mild heat stress conditions. Rather, compartment assembly also required the concerted action of molecular chaperones, protein-sorting factors and protein-sequestration factors, thus defining a minimal machinery for spatial PQC. Expression of this machinery was limited to times of acute stress through rapid changes in mRNA abundance and a proteasomal feedback mechanism. These findings demonstrate that yeast cells can control the amount of soluble misfolded proteins through regulated phase transitions in the cytoplasm, thus allowing them to rapidly adapt to changing environmental conditions.
Cellular Strategies to Counteract Protein Misfolding
Proteins misfold because of random fluctuations in protein conformations, the presence of destabilizing mutations, stress conditions, or global alterations in cellular physiology. Protein misfolding is also emerging as a major cause of human disease. Diseases that are caused or exacerbated by misfolding events include lysosomal storage disorders, cystic fibrosis, cancer and a range of neurodegenerative disorders, such as Alzheimer, Huntington or Parkinson disease. Finally, protein misfolding and aggregation are hallmarks of aging. Together, these examples illustrate that protein homeostasis is a key requirement for cellular and organismal health and survival.
Organisms have evolved an elaborate PQC machinery to preserve the functionality of their proteomes. Three different strategies are employed to counteract protein damage: refolding of misfolded proteins by molecular chaperones, removal of misfolded proteins by energy-dependent proteases or degradation by autophagy, a process of selective uptake and degradation of damaged proteins in membrane-enclosed compartments.Citation1 As a result misfolded proteins are either repaired or eliminated.
In recent years, evidence of yet another PQC mechanism has been accumulating.Citation2 Organisms as diverse as bacteria and humans have the ability to reversibly constrain misfolded proteins in specialized membrane-free compartments or inclusion bodies. Not surprisingly, the formation of these compartments is strongly enhanced by environmental stress. Inclusions are also commonly associated with protein misfolding diseases. In addition to disease-associated proteins, they can contain ubiquitylated proteins, defective ribosomal products or oxidatively damaged proteins. However, whether these proteins reside in the same compartment or are targeted to different compartments had long remained undetermined.
This situation changed in 2008 when a seminal report described two distinct stress-inducible compartments for misfolded proteins termed IPOD (insoluble protein deposit) and JUNQ (juxtanuclear quality control) in eukaryotic cells.Citation3 The juxtanuclear compartment was associated with the nuclear envelope and contained ubiquitylated proteins that rapidly exchanged with the surrounding cytosol. Additional constituents were identified as proteasomes and chaperones. Consequently, the JUNQ was proposed to provide a specialized environment for chaperone-mediated refolding or protein degradation.Citation3,Citation4 The IPOD on the other hand was identified in the perivacuolar region of yeast cells. It contained largely immobile aggregates of amyloid-forming proteins. Sequestration in the IPOD may reduce aberrant interactions between misfolded proteins and essential cellular components. This led to the proposal that the IPOD could serve a cytoprotective function.Citation3,Citation4 Despite these advances, however, the molecular mechanisms that govern the formation of aggregate deposition sites have so far remained largely elusive.
We used a yeast prion reporter assay to identify cellular factors involved in protein aggregation.Citation5 Our study identified Btn2 and Cur1, two members of the Hook family of transport factors, as potent modifiers of a synthetic yeast prion. Surprisingly, we found that the effects of Btn2 and Cur1 were not direct—as previously suggested for a different prionCitation6—but largely mediated by nuclear targeting of Sis1, a chaperone of the Hsp40 family that is required for prion maintenance. Further investigations revealed that Btn2 and Cur1 are important protein-sorting factors that physically and functionally interact with Sis1 or the small heat shock protein Hsp42 to coordinate the accumulation of aggregates in deposition sites in times of acute stress. In the following, I shall describe how these findings have advanced our understanding of spatial PQC in yeast.
Using Yeast Prions to Identify Modifiers of Protein Aggregation
Prion proteins have the remarkable ability to self-assemble, a property that is conferred by the presence of natively unfolded, amyloidogenic regions. Once a prion has formed a replication-competent structure, it undergoes a self-sustained cycle of growth and division. Hence, a critical aspect of a prion’s life cycle is the remodeling of prion polymers into heritable seeds or propagons, which can be passed from mother to daughter cells.Citation7 The remodeling reaction requires a minimal chaperone machinery consisting of Hsp40, Hsp70 and Hsp104.Citation8,Citation9 Thus, in the context of a living cell, prions and chaperones form an intricate network of physical and functional interactions that ensures faithful replication of the prion state.
The genetic tractability of S. cerevisiae has favored the discovery of a large set of prion-forming sequences in the yeast proteome.Citation10 This repertoire of diverse prion-forming sequences provides a unique opportunity to study the interactions between amyloids and molecular chaperones in the cellular milieu. Because the expression of many chaperones is subject to dramatic changes during heat shock, we investigated the effect of heat stress on the inheritance of various prions. To our surprise, we found that even mildly increased temperatures destabilized many of them (our unpublished results). The prion that showed the strongest response to heat was a synthetic construct consisting of the prion-forming domain of the RNA-binding protein Nrp1 fused to a prion reporter. Through subsequent genetic experiments we could demonstrate that prion loss was caused by upregulation of the stress-inducible protein Cur1. Btn2, a paralogue of Cur1, was also induced under heat stress, but the amount of stress-induced Btn2 was not sufficient to cause prion loss. However, Btn2 was able to destabilize the prion (termed [NRP1C+]) when overexpressed. These findings indicate that stressed yeast induce the expression of two related proteins that interfere with prion maintenance.
A first hint that pointed toward the molecular function of Btn2 and Cur1 came from the discovery that Sis1 acted as a very potent suppressor of heat-induced prion loss. Further experiments demonstrated that [NRP1C+] fibers recruit large amounts of Sis1. Because prion fiber remodeling requires the activity of Hsp40s,Citation8,Citation9 we hypothesized that Sis1-decorated prion fibers could attract Hsp104. Indeed, subsequent experiments confirmed that prion fragmentation was more pronounced in cells that overproduced Sis1. Moreover, we found that [NRP1C+] was destabilized when Sis1 was depleted. Thus, analogous to what has been proposed for other prions, maintenance of [NRP1C+] is strictly dependent on Sis1. This finding lends further support to the notion that Sis1 has an indispensable function in promoting the propagation of most, if not all yeast prions.Citation11
The functional relationship between Sis1, Btn2 and Cur1 suggested that these proteins could be interacting with each other. Co-immunoprecipitation experiments with yeast cell lysate and in vitro binding studies indeed revealed that Sis1 forms a complex with Btn2 or Cur1. From these findings, we concluded that Btn2 and Cur1 alter the activity or availability of Sis1.
Btn2 and Cur1 Alter the Subcellular Localization of Sis1
Using a phenotypic assay for a synthetic yeast prion, we were able to identify Btn2 and Cur1 as interactors and functional modifiers of Sis1. To investigate the functional significance of this interaction, we first explored the behavior of GFP-tagged Sis1 in yeast cells that were subject to a mild temperature increase. In stressed cells, Sis1 accumulated in the nucleus and in peripheral and juxtanuclear puncta. Remarkably, we observed very similar patterns in unstressed cells that overexpressed Btn2 and Cur1. Furthermore, localization of Sis1 to the nucleus and to cytosolic compartments was strongly reduced in stressed cells that lacked Btn2 and/or Cur1. Thus, we concluded that Btn2 and Cur1 determine the subcellular distribution of Sis1 during acute heat stress.
Nuclear accumulation of Sis1 was dependent on Btn2 and Cur1, suggesting that Btn2 and Cur1 could be nuclear targeting factors for Sis1. Indeed, we were able to identify classical nuclear localization signals in Btn2 and Cur1. Mutants lacking the NLS motif showed diminished targeting to the nucleus and to the juxtanuclear compartment. The mutant proteins were also no longer able to enrich Sis1 in the nucleus. Further experiments revealed that complex formation between Sis1 and Btn2 or Cur1 was a prerequisite for sorting to the nucleus. Moreover, we found that nuclear targeting was dependent on Srp1, the only member of the α-importin family in yeast. Thus, sorting of Sis1 to the nucleus requires nuclear localization sequences in Btn2 or Cur1 and the α-importin Srp1.
Aggregate Deposition Requires Btn2, Sis1 and Hsp42
In stressed yeast cells, Sis1 not only accumulated in the nucleus but also coalesced into foci, which were reminiscent of previously described PQC compartments. To determine whether Sis1 is sorted to these compartments, we used the misfolding-prone marker proteins VHL and Ubc9ts.Citation3 Indeed, we found that Sis1 showed substantial overlap with both aggregated VHL and Ubc9ts. In addition, Sis1-positive sites contained various chaperones such as Hsp42, Ssa1 and Hsp104. Thus, during acute stress, Sis1 accumulates in cytosolic deposition sites that contain aggregation-prone proteins and components of the PQC machinery.
A property that distinguished Btn2 from Cur1 was that Btn2 strongly localized to a peripheral protein deposition site. Given the central role of Hsp42 in controlling the formation of this compartment,Citation12 we hypothesized that Btn2 could be accumulating in the peripheral site in an Hsp42-dependent manner. Indeed, we found that Btn2 and Hsp42 formed a complex in the periphery. In cells that were eliminated for Hsp42, Btn2 no longer accumulated in peripheral deposition sites. This indicates that association of Btn2 and Hsp42 is a prerequisite for accumulation of Btn2 in the perivacuolar region.
Collectively, these data suggested a role for Btn2 and Cur1 in aggregate sorting. To investigate this possibility, we performed a time-resolved analysis of VHL aggregation in cells that were eliminated for Btn2 and/or Cur1. In Btn2-deficient cells, VHL was no longer sorted to juxtanuclear sites, and the size of peripheral sites was strongly reduced. Remarkably, we found that overexpression of Btn2 alone was sufficient to induce the formation of aggregate deposition sites. Additional experiments revealed that association of Btn2 with Sis1 or Hsp42−where the two chaperones serve as adaptors for the recognition of misfolded proteins−is the key molecular event that determines whether a misfolded protein is sorted to a juxtanuclear or a peripheral site. In contrast to Btn2, we could not, however, find evidence for a direct role of Cur1 in aggregate sorting.
The Molecular Mechanism of Prion Curing by Btn2 and Cur1
In the course of our studies, we noticed that the nuclear targeting potential of Btn2 and Cur1 was very different: while Btn2 caused only a slight nuclear enrichment of Sis1, Cur1 expression led to almost complete nuclear accumulation. Given that Btn2 also had a lower prion-curing capacity than Cur1, this suggested that cytosolic depletion of Sis1 could be the underlying mechanism of prion loss. To test this assumption, we generated a variant of Sis1 (NLS-Sis1) that was able to target endogenous Sis1 to the nucleus. Overexpression of NLS-Sis1 indeed led to destabilization of [NRP1C+], suggesting that redistribution of Sis1 to the nucleus is sufficient to induce prion loss. Additional genetic experiments showed that NLS-deficient Btn2 and Cur1 had a reduced prion-curing potential. Thus, by targeting Sis1 to the nucleus, Btn2 and Cur1 limit the amount of Sis1 that is available for prion binding and fragmentation. Reduced prion fragmentation rates, however, cause a net increase in prion polymer length. This could eventually lead to prion loss in populations of dividing yeast cells. Consistent with this hypothesis, Btn2- and Cur1-dependent prion loss was found to require cell division.Citation6 Moreover, we discovered that prion polymers were significantly shorter in cells that overexpressed Sis1 or lacked Btn2 or Cur1. Based on these findings, we concluded that Btn2 and Cur1 interfere with prion propagation through depletion of Sis1 from the cytosol.
A previous study proposed an alternative mechanistic explanation for prion curing by Btn2.Citation6 According to this view, Btn2 directly promotes the sequestration of prion aggregates and could potentially induce a pathway that facilitates the disposal of prions by autophagy. Consistent with such a direct role in prion sequestration, Btn2 was shown to colocalize with prions.Citation6,Citation13 However, using improved imaging technology, we could demonstrate that the spatial overlap between Btn2 and prions was very limited. In agreement with our finding, Kryndushkin et al. (2008) could not detect a direct interaction between purified Btn2 and prion proteins in an in vitro binding assay.Citation6 Moreover, Btn2 had no effect on the assembly kinetics of Ure2. Therefore, current evidence is arguing against a direct role of Btn2 in prion sequestration.
Although a direct role of Btn2 in prion sequestration is unlikely, Btn2 could still cooperate with other factors such as Hsp42 to promote the sequestration of misfolded prion proteins in a peripheral location. In agreement with this idea, Hsp42 has recently been shown to interfere with prion inheritance by binding to prion fibers.Citation14 Therefore, aggregated prion proteins that associate with Hsp42 could in principle be targeted to a peripheral deposition site in a Btn2-dependent manner. However, cells that lacked Btn2 and/or Hsp42 showed no discernable defects in localizing prion aggregates to a peripheral location (Specht et al. and our unpublished observations).Citation12 This suggests that prion targeting to the peripheral compartment is largely independent of Btn2 and Hsp42. These findings and considerations further reinforce our conclusion that nuclear targeting of Sis1 is the predominant mechanism of prion curing by Btn2 and Cur1. Future experiments with Btn2 and Cur1 mutants carrying specific Sis1-binding defects are required to determine whether prion loss is entirely due to Sis1 depletion or might involve other molecular mechanisms.
Btn2 and Cur1 are Stress-Inducible and Short-Lived
Juxtanuclear and peripheral aggregate deposition sites are hardly detectable in cells growing under normal conditions, but strongly enlarged when cells experience acute stress. In line with this observation, the transcripts of BTN2 and CUR1 are upregulated in yeast that are exposed to different stresses such as ethanol, high salt concentrations or heat.Citation15-Citation18 In addition, Btn2 is strongly induced in cells that overexpress proteotoxic proteins.Citation19,Citation20 Consistent with this, the promoter regions of BTN2 and CUR1 were found to contain heat shock sequence elements, similar to those in other stress-inducible genes.Citation6 Collectively, these findings suggested that Btn2 and Cur1 could be involved in the environmental stress response. To determine whether Btn2 and Cur1 are induced in times of acute stress, we compared their protein levels in unstressed and stressed yeast. Indeed, Cur1 and Btn2 expression was strictly limited to times of stress. This was not only due to changes in gene expression, but also resulted from the fact that Btn2 and Cur1 were rapidly turned over by the proteasome.
Acute heat stress causes a rapid accumulation of insoluble ubiquitylated proteins.Citation21 Hence, we reasoned that a sudden buildup of substrate proteins could interfere with proteasomal degradation, causing a slower turnover of Btn2 and Cur1. To determine whether stress affects the half-life of Btn2 and Cur1, we expressed them from a non-inducible promoter. Remarkably, under these conditions Btn2 and Cur1 were strongly stabilized. This indicates the presence of a dynamic feedback mechanism that couples induction of Btn2 and Cur1 to the activity of the proteasome. Thus, changes in gene expression and a transient decrease in overall proteasomal activity jointly restrict Btn2 and Cur1 expression to periods of acute stress.
Genome-wide screens have identified many genetic modifiers of prion induction and inheritance.Citation22,Citation23 Among these positive hits, genes associated with the PQC machinery or stress response pathways were particularly prominent. For example, a deletion of the gene for the non-essential proteasome subunit Pre9 had a very pronounced inhibitory effect on prion induction.Citation22,Citation23 Remarkably, we found that the expression levels of Btn2 and Cur1 were strongly increased in Pre9-deficient cells (unpublished results). Chronic induction of short-lived sorting factors such as Btn2, Cur1 or Lsb2Citation24 could therefore provide a plausible explanation for the frequent discovery of PQC-related modifiers in prion screens. Thus, stress conditions are not only able to trigger the birth of prions but they can also cause their demise.
A Model for Spatial Protein Quality Control in Yeast
Our previous experiments revealed an important function for Btn2 in aggregate sorting. Unexpectedly, however, we found that the paralogue Cur1 did not share this ability. To investigate the function of Cur1, we performed a series of genetic experiments. In these experiments Cur1 overexpression had a strongly detrimental effect on the growth of yeast cells. This growth defect was completely reversed by co-expression of Sis1 or deletion of the nuclear localization sequence. We therefore concluded that the main function of Cur1 is to control the cytosolic availability of Sis1.
On the basis of our collective findings, we proposed the following model to describe stress-induced alterations in the subcellular distribution of misfolded proteins (). When yeast cells are exposed to heat stress, they first form a juxtanuclear site, while the formation of the peripheral compartment is delayed. These changes are predominantly determined by temporal changes in the relative cytosolic concentrations of Sis1 and Hsp42. At the onset of a stress stimulus, relative Sis1 levels are high. Under these conditions, Btn2 associates with Sis1 to target misfolded proteins to the juxtanuclear site. However, over time induction of Cur1 leads to a depletion of Sis1 from the cytosol. As a consequence, substrate flux to the juxtanuclear compartment is gradually reduced. At the same time, however, aggregate transport to the periphery increases through upregulation of Hsp42. Thus, temporal changes in the cytosolic concentration of chaperones and protein-sorting factors determine whether aggregated proteins are stored in the periphery, or shuttled to a juxtanuclear site, most likely to facilitate their refolding or degradation.
Figure 1. A model for the coordinated action of sorting factors and molecular chaperones during acute heat stress. (A) Yeast cells growing under normal conditions, (B) cells exposed to acute heat stress and (C) cells that are recovering from stress. The events that induce sorting (association with a factor or assembly into an amyloid structure) are indicated in red. A bound misfolded protein is depicted in blue. Please note that only the complexes containing Btn2 are involved in aggregate sorting, while the Cur1 complex mediates sorting of Sis1 to the nucleus. See text for more details.
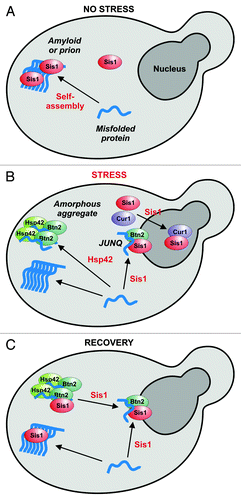
What happens when the temperature returns to normal levels? In the recovery phase, aggregate dissolution is facilitated by Hsp104.Citation12 Hsp104 is most likely assisted by members of the Hsp70 and Hsp40 families, which play an important role in providing Hsp104 with access to substrate proteins.Citation8,Citation9 Therefore, when Sis1 reenters the cytosol, it could engage with substrate proteins to promote their release from the disintegrating peripheral compartment. Association with Btn2 could then promote their targeting to the juxtanuclear site.
Surprisingly, despite their important role in regulating the aggregation of proteins, Hsp42, Btn2 and Cur1 are not essential for thermotolerance (Haslbeck et al. 2004 and our unpublished observations).Citation25 This suggests that the spatial PQC machinery has additional functions that have yet to be uncovered. Interestingly, Btn2 was initially reported to be involved in vesicular trafficking and late Golgi-endosome sorting.Citation13,Citation26,Citation27 In agreement with such a role, Btn2 was found to restore intracellular amino acid levels and vacuolar functionality in a yeast model for Batten disease, a lysosomal storage disorder.Citation15,Citation28 These findings suggest that the spatial PQC machinery could have additional functions that go beyond its role in protein quality control. Additional studies will be required to explore this exciting possibility.
Conclusion
Protein aggregation was long considered to be a chaotic process that is only initiated when the capacities of the cellular PQC machinery are exhausted. Recent evidence, however, suggests that protein aggregation can also be the result of an elaborate cellular program. Our study has provided insight into the molecular mechanisms that determine the spatiotemporal distribution of protein aggregates and their sequestration in membrane-free compartments during mild heat stress. Related protein bodies such as RNA-containing granules might use very similar mechanisms to form. Thus, a new theme is emerging that suggests that environmentally challenged cells can compartmentalize the cytoplasm through controlled phase transitions of vast numbers of individual macromolecules.
| Abbreviations: | ||
| PQC | = | protein quality control |
| NLS | = | nuclear localization sequence |
| IPOD | = | insoluble protein deposit |
| JUNQ | = | juxtanuclear protein quality control |
Acknowledgments
I am grateful to the members of the Alberti lab for critical comments on the manuscript and indebted to the Max Planck Society for funding.
References
- Buchberger A, Bukau B, Sommer T. Protein quality control in the cytosol and the endoplasmic reticulum: brothers in arms. Mol Cell 2010; 40:238 - 52; http://dx.doi.org/10.1016/j.molcel.2010.10.001; PMID: 20965419
- Tyedmers J, Mogk A, Bukau B. Cellular strategies for controlling protein aggregation. Nat Rev Mol Cell Biol 2010; 11:777 - 88; http://dx.doi.org/10.1038/nrm2993; PMID: 20944667
- Kaganovich D, Kopito R, Frydman J. Misfolded proteins partition between two distinct quality control compartments. Nature 2008; 454:1088 - 95; http://dx.doi.org/10.1038/nature07195; PMID: 18756251
- Chen B, Retzlaff M, Roos T, Frydman J. Cellular strategies of protein quality control. Cold Spring Harb Perspect Biol 2011; 3:a004374; http://dx.doi.org/10.1101/cshperspect.a004374; PMID: 21746797
- Malinovska L, Kroschwald S, Munder MC, Richter D, Alberti S. Molecular chaperones and stress-inducible protein-sorting factors coordinate the spatiotemporal distribution of protein aggregates. Mol Biol Cell 2012; 23:3041 - 56; http://dx.doi.org/10.1091/mbc.E12-03-0194; PMID: 22718905
- Kryndushkin DS, Shewmaker F, Wickner RB. Curing of the [URE3] prion by Btn2p, a Batten disease-related protein. EMBO J 2008; 27:2725 - 35; http://dx.doi.org/10.1038/emboj.2008.198; PMID: 18833194
- Tuite MF, Serio TR. The prion hypothesis: from biological anomaly to basic regulatory mechanism. Nat Rev Mol Cell Biol 2010; 11:823 - 33; http://dx.doi.org/10.1038/nrm3007; PMID: 21081963
- Tipton KA, Verges KJ, Weissman JS. In vivo monitoring of the prion replication cycle reveals a critical role for Sis1 in delivering substrates to Hsp104. Mol Cell 2008; 32:584 - 91; http://dx.doi.org/10.1016/j.molcel.2008.11.003; PMID: 19026788
- Winkler J, Tyedmers J, Bukau B, Mogk A. Hsp70 targets Hsp100 chaperones to substrates for protein disaggregation and prion fragmentation. J Cell Biol 2012; 198:387 - 404; http://dx.doi.org/10.1083/jcb.201201074; PMID: 22869599
- Alberti S, Halfmann R, King O, Kapila A, Lindquist S. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell 2009; 137:146 - 58; http://dx.doi.org/10.1016/j.cell.2009.02.044; PMID: 19345193
- Higurashi T, Hines JK, Sahi C, Aron R, Craig EA. Specificity of the J-protein Sis1 in the propagation of 3 yeast prions. Proc Natl Acad Sci U S A 2008; 105:16596 - 601; http://dx.doi.org/10.1073/pnas.0808934105; PMID: 18955697
- Specht S, Miller SB, Mogk A, Bukau B. Hsp42 is required for sequestration of protein aggregates into deposition sites in Saccharomyces cerevisiae. J Cell Biol 2011; 195:617 - 29; http://dx.doi.org/10.1083/jcb.201106037; PMID: 22065637
- Kanneganti V, Kama R, Gerst JE. Btn3 is a negative regulator of Btn2-mediated endosomal protein trafficking and prion curing in yeast. Mol Biol Cell 2011; 22:1648 - 63; http://dx.doi.org/10.1091/mbc.E10-11-0878; PMID: 21441304
- Duennwald ML, Echeverria A, Shorter J. Small heat shock proteins potentiate amyloid dissolution by protein disaggregases from yeast and humans. PLoS Biol 2012; 10:e1001346; http://dx.doi.org/10.1371/journal.pbio.1001346; PMID: 22723742
- Chattopadhyay S, Muzaffar NE, Sherman F, Pearce DA. The yeast model for batten disease: mutations in BTN1, BTN2, and HSP30 alter pH homeostasis. J Bacteriol 2000; 182:6418 - 23; http://dx.doi.org/10.1128/JB.182.22.6418-6423.2000; PMID: 11053386
- Espinazo-Romeu M, Cantoral JM, Matallana E, Aranda A. Btn2p is involved in ethanol tolerance and biofilm formation in flor yeast. FEMS Yeast Res 2008; 8:1127 - 36; http://dx.doi.org/10.1111/j.1567-1364.2008.00397.x; PMID: 18554307
- Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, et al. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell 2000; 11:4241 - 57; PMID: 11102521
- Melamed D, Pnueli L, Arava Y. Yeast translational response to high salinity: global analysis reveals regulation at multiple levels. RNA 2008; 14:1337 - 51; http://dx.doi.org/10.1261/rna.864908; PMID: 18495938
- Hughes RE, Lo RS, Davis C, Strand AD, Neal CL, Olson JM, et al. Altered transcription in yeast expressing expanded polyglutamine. Proc Natl Acad Sci U S A 2001; 98:13201 - 6; http://dx.doi.org/10.1073/pnas.191498198; PMID: 11687606
- Treusch S, Lindquist S. An intrinsically disordered yeast prion arrests the cell cycle by sequestering a spindle pole body component. J Cell Biol 2012; 197:369 - 79; http://dx.doi.org/10.1083/jcb.201108146; PMID: 22529103
- Fang NN, Ng AH, Measday V, Mayor T. Hul5 HECT ubiquitin ligase plays a major role in the ubiquitylation and turnover of cytosolic misfolded proteins. Nat Cell Biol 2011; 13:1344 - 52; http://dx.doi.org/10.1038/ncb2343; PMID: 21983566
- Manogaran AL, Hong JY, Hufana J, Tyedmers J, Lindquist S, Liebman SW. Prion formation and polyglutamine aggregation are controlled by two classes of genes. PLoS Genet 2011; 7:e1001386; http://dx.doi.org/10.1371/journal.pgen.1001386; PMID: 21625618
- Tyedmers J, Madariaga ML, Lindquist S. Prion switching in response to environmental stress. PLoS Biol 2008; 6:e294; http://dx.doi.org/10.1371/journal.pbio.0060294; PMID: 19067491
- Chernova TA, Romanyuk AV, Karpova TS, Shanks JR, Ali M, Moffatt N, et al. Prion induction by the short-lived, stress-induced protein Lsb2 is regulated by ubiquitination and association with the actin cytoskeleton. Mol Cell 2011; 43:242 - 52; http://dx.doi.org/10.1016/j.molcel.2011.07.001; PMID: 21777813
- Haslbeck M, Braun N, Stromer T, Richter B, Model N, Weinkauf S, et al. Hsp42 is the general small heat shock protein in the cytosol of Saccharomyces cerevisiae. EMBO J 2004; 23:638 - 49; http://dx.doi.org/10.1038/sj.emboj.7600080; PMID: 14749732
- Chattopadhyay S, Roberts PM, Pearce DA. The yeast model for Batten disease: a role for Btn2p in the trafficking of the Golgi-associated vesicular targeting protein, Yif1p. Biochem Biophys Res Commun 2003; 302:534 - 8; http://dx.doi.org/10.1016/S0006-291X(03)00209-2; PMID: 12615067
- Kama R, Robinson M, Gerst JE. Btn2, a Hook1 ortholog and potential Batten disease-related protein, mediates late endosome-Golgi protein sorting in yeast. Mol Cell Biol 2007; 27:605 - 21; http://dx.doi.org/10.1128/MCB.00699-06; PMID: 17101785
- Kim Y, Chattopadhyay S, Locke S, Pearce DA. Interaction among Btn1p, Btn2p, and Ist2p reveals potential interplay among the vacuole, amino acid levels, and ion homeostasis in the yeast Saccharomyces cerevisiae. Eukaryot Cell 2005; 4:281 - 8; http://dx.doi.org/10.1128/EC.4.2.281-288.2005; PMID: 15701790