Abstract
The pH-dependence of the ability of Bgl2p to form fibrils was studied using synthetic peptides with potential amyloidogenic determinants (PADs) predicted in the Bgl2p sequence. Three PADs, FTIFVGV, SWNVLVA and NAFS, were selected on the basis of combination of computational algorithms. Peptides AEGFTIFVGV, VDSWNVLVAG and VMANAFSYWQ, containing these PADs, were synthesized. It was demonstrated that these peptides had an ability to fibrillate at pH values from 3.2 to 5.0. The PAD-containing peptides, except for VDSWNVLVAG, could fibrillate also at pH values from pH 5.0 to 7.6. We supposed that the ability of Bgl2p to form fibrils most likely depended on the coordination of fibrillation activity of the PAD-containing areas and Bgl2p could fibrillate at mild acid and neutral pH values and lose the ability to fibrillate with the increasing of pH values. It was demonstrated that Bgl2p was able to fibrillate at pH value 5.0, to form fibrils of various morphology at neutral pH values and lost the fibrillation ability at pH value 7.6. The results obtained allowed us to suggest a new simple approach for the isolation of Bgl2p from Saccharomyces cerevisiae cell wall.
Introduction
Amyloids are fibrillar aggregates of proteins consisting of long β-sheets in which β-strands are located perpendicular to the fibril axis (cross-β-structure).Citation1 Initially, amyloids attracted researchers’ attention because of the fact that many human and animal diseases, including Alzheimer and Parkinson disease, are associated with formation of amyloid deposits called “amyloid plaques” in various tissues.Citation2 However, over the last decade a diversity of proteins have been shown to fulfill their role in vivo in the form of amyloid fibrils.Citation3-Citation6 Amyloids were found to be highly widespread at the cell surface of various microorganisms, including fungi.Citation7-Citation11 Hydrophobins,Citation3Citation9 repellents,Citation10 and adhesinsCitation11 have been shown to form amyloids on fungal surfaces and many secreted cell wall proteins of Ustilago maydis are predicted to be amyloids.Citation12 The investigation of amyloid proteins of microorganisms is of special importance for understanding of the potential of microbe amyloids to cause harm to human and animal health, particularly by constituting the “nucleus” of amyloid deposits in macroorganisms.Citation13
Recently, we studied the cell wall (CW) of Saccharomyces cerevisiae yeast, a microorganism of great significance for industry, medicine and pharmacology, in order to identify amyloid-forming proteins. We showed that the major, conserved and thermostable cell wall proteinCitation14,Citation15 glucantransferase Bgl2p, described for a wide range of yeast species, formed the amyloid aggregates.Citation16 The amyloid-like characteristics of Bgl2p include fibrillar morphology of the aggregates revealed in Bgl2p preparations using transmission electron microscopy, as well as interaction of Bgl2p-containing CW with Congo Red (CR), giving strong green birefringence, and β-sheet-rich secondary structure shown by circular dichroism analysis and thioflavin T (ThT) fluorescence.Citation16 Whether there were conditions, in which Bgl2p did not show the ability to fibrillate, had been unknown by the time we started this work.
In view of the importance of understanding the mechanism of amyloid fibril formation for biotechnology, biology and medicine, it is necessary to investigate factors and conditions which define the existence of proteins in a soluble or an amyloid form.Citation17 Also it was suggested that the key elements of the fibril formation may be common to different proteins and simple model systems could help to clarify many general aspects of this process.Citation18 In the present work, the Bgl2p fibrillation process was investigated more thoroughly using Bgl2p synthetic peptides as model systems. We focused on the pH dependence of the fibrillation processes because of the facts that the pH value has a strong impact on the amyloid fibrillation tendency and the structure of amyloid fibrils depends on the pH at which they are prepared.Citation18,Citation19
This approach allowed us to identify trends in the ability of Bgl2p, isolated from cell walls by heating, to fibrillate at different pH values, based on which we offered a new method of Bgl2p extraction from yeast CW.
Results
Bioinformatic analysis of potential amyloidogenic determinants in Bgl2p
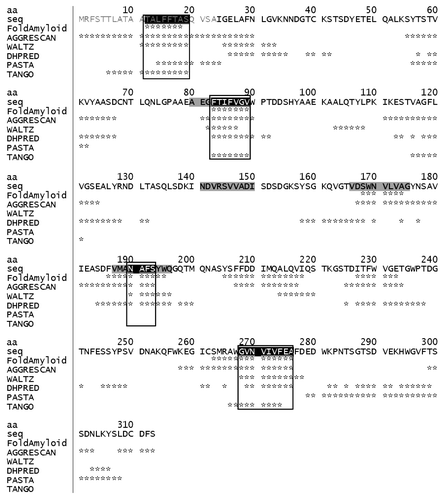
The Bgl2p sequence was analyzed using six computational algorithms (FoldAmyloid, TANGO, AGGRESCAN, PASTA, WALTZ and DHPRED), and the obtained results were compared. The underlying principles of these algorithms can be found in the articles dedicated to these issues.Citation20-Citation28 The results of the analysis are presented in . Several potential amyloidogenic determinants (PADs) were predicted at least by four or even by five methods out of six. The predicted PADs were TALFFTAS (аа 12-19), FTIFVGV (аа 83-89), NAFS (аа 190-193) and GVNVIVFEA (аа 268-276). The rest of the protein sequence was presented by areas, which none of the programs used predicted as potentially amyloid ones, as well as by those that were predicted to be potentially amyloid, but by less than four of the programs. Several sequences were predicted by FoldAmyloid and AGGRESCAN. It should be noted that the comparison of predicted aggregation propensity results by different methods and experimental data obtained in vivo the programs FoldAmyloid and AGGRESCAN demonstrated better results than in the case of the TANGO, PASTA and WALTZ algorithms.Citation17 To verify whether the predicted PADs of Bgl2p really had the pronounced propensity to form amyloid structures, we synthesized PAD-containing peptides with a length of 10 aa.
Figure 1. Potential amyloidogenic determinants in Saccharomyces cerevisiae cell wall glucantransferase Bgl2p (UniProtKB/TrEMBL entry number P15703). The amino acids predicted by computational algorithms to be a part of potential amyloidogenic determinants are marked with (*) opposite the name of the corresponding algorithm. The confluences of positive amyloidogenic determinant prediction results are surrounded by frames. Bgl2p N-terminal cell wall transport signal (in gray letters); peptide sequences, which were synthesized and investigated, are saturated gray. Abbreviations: aa, amino acid number; seq, amino acid sequence.
Synthesis of peptides
The peptides were synthesized with blocked (acetylated and amidated) and non-blocked termini. Hereinafter “NB” and “B” at the end of peptides mean “non-blocked” and “blocked” termini, respectively. The first predicted PAD (аa 12-19) was discarded, since it fell into the Bgl2p post-translationally processed N-terminal cell wall transport signal region, which was absent in Bgl2p from CW. Unfortunately, the peptides which contained PAD (aa 268-276) turned out to be insoluble in all solvents tested, therefore this PAD was also not studied further. The control peptide NDVRSVVADI (aa 141-150 “B”) from the region which was predicted to be non-amyloid according to our bioinformatic analysis, was synthesized as well. Because several sequences were predicted by FoldAmyloid and AGGRESCAN, we decided to synthesize peptides VDSWNVLVAG (aa 166-175 “B” and “NB”) with one of such sequences, SWNVLVA (aa 168-174).
In further experiments the soluble PAD-containing peptides AEGFTIFVGV (aa 80-89 “B”) and VMANAFSYWQ (aa 187-196 “B” and “NB”), the peptides VDSWNVLVAG (aa 166-175 “B” and “NB”) and the non-amyloidogenic control peptide NDVRSVVADI (aa 141-150 “B”) were examined.
Effect of pH value on fibrillation of peptides
In the first step of our work the ability of the peptides to form fibrils was determined using the amyloid-specific fluorescent probe ThT.Citation29 To avoid presence of charges on termini of the peptides investigated (to keep their state close to the one inside the Bgl2p molecule) measurements of ThT fluorescence of peptides with blocked (acetylated and amidated) termini were made. Peptides AEGFTIFVGV (aa 80-89 “B”) and VMANAFSYWQ (aa 187-196 “B”), containing PADs of different length, fibrillated at all pH values investigated (3.2–7.6) after half an hour of shaking (). Peptide VDSWNVLVAG (aa 166-175 “B”) did not fibrillate during this time frame (half an hour), but it fibrillated at pH 3.2, 3.9 and 4.5 after shaking during 1.0, 1.5 and 14.0 h consequently (). Fibrils of this peptide at pH value 5.0 were visualized with ThT using fluorescent microscopy (). Thus, peptide VDSWNVLVAG (aa 166-175 “B”) fibrillated at pH values from 3.2 to 5.0 and it can also be viewed as а PAD-containing peptide. At pH ranging from 6.2 to 7.6 the fibrillation of this peptide was not detected (data not shown). The non-amyloidogenic control peptide NDVRSVVADI (aa 141-150 “B”) did not induce the characteristic ThT fluorescence at pH values ranging from 3.2 to 7.6 even after shaking for 14 h (data not shown).
Table 1. Amyloid formation ability of the Bgl2p PAD-containing peptides (aa 80-89 “B”) and (aa 187-196 “B”)
Table 2. Amyloid formation ability of the Bgl2p PAD-containing peptide (aa 166-175 “B”)
Figure 2. Fluorescence microscopy of the peptides at pH 5.0. (A) The peptide (aa 166-175 “B”). (B) The peptide (aa 166-175 “NB”). Staining was done with 7.5 μM ThT.
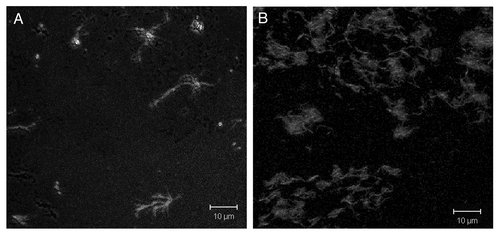
Fibrils of peptide VDSWNVLVAG (aa 166-175 “NB”) at pH value 5.0 were visualized with ThT in microscopy (). Some additional experimental data in support of the amyloid properties of the investigated peptides with PADs is given in Supplementary Information section (Figs. S1–S3 and Table S1).
Fibril forming ability of Bgl2p
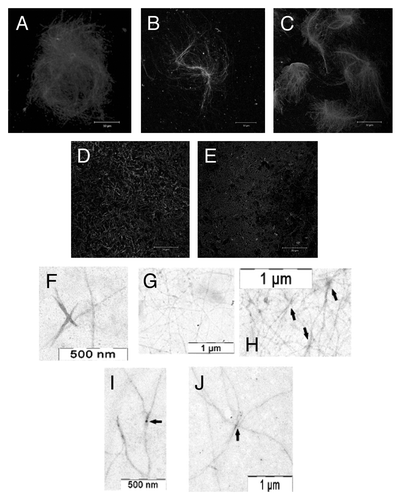
The fibrillation kinetics of Bgl2p was studied at pH 5.0 and 7.6 using ThT. Bgl2p induced specific ThT fluorescence at pH 5.0, but did not induce specific ThT fluorescence at pH 7.6 (). The ability of Bgl2p to form fibrils in water was also demonstrated (pH average 6.4 ± 0.2). Using microscopy it was demonstrated that Bgl2p isolated from the CW without trypsin treatment formed long (up to several tens of µm) fibrils and aggregates resembling “wisps” or “nests” with evident aggregation centers (Fig. Four A-C). When Bgl2p was isolated from the trypsin-treated CW, the fibrils detected were curved, “worm-like” and rather short (up to 10 μm) (). In this case only a few evident aggregation centers were revealed only by TEM (, arrows). Addition of the major CW component, β-1,3-glucan (laminarin), did not affect the morphology and amount of Bgl2p fibrils (data not shown). However, electron microscopy revealed clear aggregation centers in presence of β-1,3-glucan (). Notably, the common feature of all fibrils was their length of more than 1 μm.
Figure 3. Fibrillation kinetics of Bgl2p at different pH values measured by thioflavin T fluorescence at 480 nm at pH 5.0 (solid line) and pH 7.6 (dashed line). The fibrillation kinetics was measured at 35°C and using 600 rpm of shaking for 240 sec during each cycle of 400 sec. The excitation wavelength was 450 nm. The plotted data curves are an average of the three individually measured fibrillation kinetics.
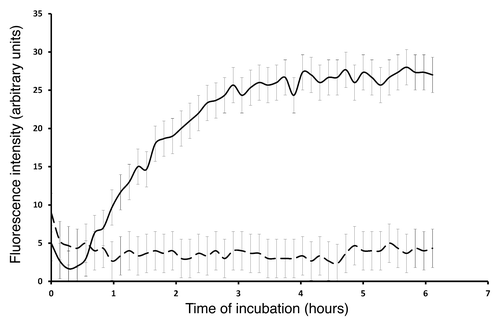
Figure 4. Bgl2p fibril morphology. (A–E) Immunofluorescent microscopy using anti-Bgl2p antibody. Bgl2p was isolated from the CW, which was either not treated (A–C) or treated (D and E) with trypsin before Bgl2p extraction from Saccharomyces cerevisiae cell wall. (F– J) Transmission electron microscopy. Bgl2p was isolated from trypsin-treated Saccharomyces cerevisiae cell wall and then it was incubated in absence (F–H) or in presence (I and J) of laminarin. Arrows indicate the “aggregation centers.”
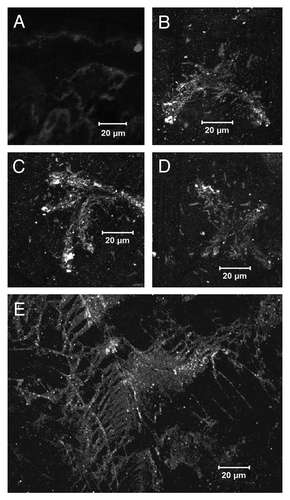
Bgl2p, which was partially purified by ultrafiltration (cut off limit 100 kDa) from the growth medium of mutant strain S. cerevisiae A270 with deletion in SSU/MCD4 gene (Bgl2pGM), at рН 6.7 formed unstructured aggregates in the absence () and fibrillar structures in presence of CR (). CR is known as an agent, which can either inhibit or enhance fibrillation.Citation29,Citation30 Fibrillar structures were also found in growth media subjected to dialysis and partial concentration by ultrafiltration in order to eliminate of components with molecular mass less than 10 kDa () without addition of CR.
Figure 5. Bgl2pGM fibril morphology. Immunofluorescent microscopy of Bgl2pGM fibrils with anti-Bgl2p antibody. Ultrafiltered growth medium of Saccharomyces cerevisiae ssu21/mcd4 mutant contains components which pass through the membrane with a cut-off limit of 100 kDa. Images of these components were collected without Congo red (A) and in presence of 8 µM Congo red (B–D). The growth medium containing components which do not pass through the membrane with a cut-off limit of 10 kDa (E).
According to the results presented in the Supplementary Information (Fig. S4), no difference was observed between the mutant growth medium Bgl2p (Bgl2pGM) and wild-type CW Bgl2p in their primary structure and in post-translational modifications. It is important to note as well that Bgl2pGM was able to form denaturation-resistant oligomers (Fig. S5) and CW Bgl2p demonstrated proteinase K resistance (Fig. S6).
Isolation of Bgl2p from cell walls without heating
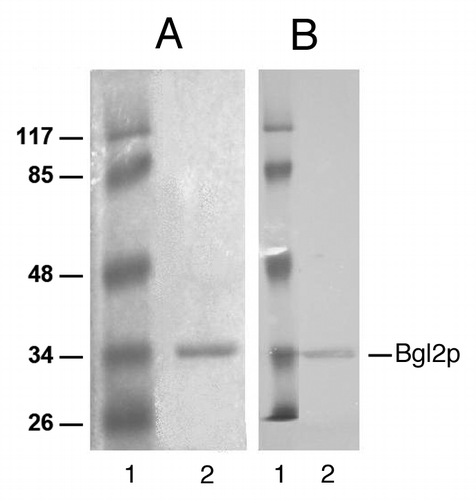
Previously, we suggested that the ability to form amyloid-like fibrils allows Bgl2p to be retained firmly in the cell wall.Citation16 In this work we demonstrated that the fibrillation ability of this protein extracted from a cell wall by heating was decreasing with the increase of pH value. We took into consideration such pH dependence tendency of Bgl2p fibrillation and extracted Bgl2p by incubation of cell walls in a solution with alkaline pH (). It was possible to extract Bgl2p at a pH value between 8 and 9, but the extraction was either slow (in this case the cell walls had to be incubated at alkaline pH for more than 24 h) or ineffective (data not shown). At pH 9 efficient extraction conditions were shown, since in a relatively short time period the amount of Bgl2p comparable to the amount of the protein extracted from the cell walls by heating was extracted.
Discussion
Bgl2p has been studied for a long time but only recently it has been shown that this protein possesses amyloid properties.Citation16 However, conditions influencing on Bgl2p fibrillation have not been studied until now.
According to the literature, one of the important factors that affects the fibrillation is pH value of the medium.Citation18,Citation19 Therefore we focused on studying the pH dependence of Bgl2p fibrillation. The pH-dependence of the ability of Bgl2p to form fibrils was studied using synthetic peptides with potential amyloidogenic determinants (PADs) predicted in the Bgl2p sequence according to a bioinformatic approach.
Structural studies as well as in vitro experiments with proteolytic fragments of amyloidogenic proteins and synthetic peptides revealed that the tendency for a protein to form amyloid is often limited to a short sequence of a full protein, known as a “self-recognition element.”Citation31 Self-recognition elements constitute the core of amyloid fibrils,Citation32,Citation33 and “hot spots” for aggregation of the native protein into amyloid fibrils.Citation34 In this article, such self-recognition elements are referred to as “potential amyloidogenic determinants” (PADs).
In this work Bgl2p peptide sequences, determined to be important for the amyloidogenic properties of Bgl2p using various bioinformatics analyses, were validated experimentally mainly using the ThT assay. The selected PAD sequences were predicted to be PADs by at least four out of six prediction algorithms. Furthermore, a sequence only selected by FoldAmyloid and AGGRESCAN was also selected, because these two programs had earlier demonstrated a high level of correlation between the change in aggregation propensity observed in vivo and the change in aggregation propensity predicted in silico.Citation17
Our prediction of amyloidogenic determinants in the Bgl2p sequence correlated well with the data obtained on the fibrillation ability of the peptides ( and and ). The PAD-containing peptides AEGFTIFVGV (aa 80-89 “B”) and VMANAFSYWQ (aa 187-196 “B”) formed amyloids at all pH values investigated (), the peptide NDVRSVVADI (aa 141-150 “B”), which was predicted to be non-amyloidogenic, did not form amyloids (data not shown). The peptides VDSWNVLVAG (aa 166-175 “B” and “NB”) also formed amyloids within the pH range 3.2–5.0 ( and ). For comparison some additional experiments revealing the amyloid properties of the peptides investigated (with non-blocked termini) are presented in Supplementary Information (Figs. S1–S3 and Table S1). These experiments supported the presence of amyloid-like aggregates in the samples of peptides that showed a positive ThT signal.
Bgl2p had the same tendency to fibrillate as the peptides VDSWNVLVAG (aa 166-175 “B” and “NB”) ( and ): it fibrillated at pH 5.0 and did not fibrillate at mild alkaline pH value 7.6. Thus, apparently the Bgl2p ability to fibrillate at different pH-values was the function of coordinated fibrillation activity of different peptides.
The new insight into the pH-dependence of Bgl2p fibrillation was used to introduce a novel extraction method where Bgl2p was extracted from the cell wall without heating. It is advantageous to avoid heating of Bgl2p although Bgl2p is considered to be a thermally stable protein, since its activity when using the heating method of extraction is rather low,Citation35 which may in some cases lead to misinterpretation of the results.Citation14,Citation15 Furthermore, earlier the isolation of Bgl2p without heating was an extremely complicated and laborious procedure.Citation15 As follows from the results obtained, extraction of Bgl2p at alkaline pH from the cell walls allowed us to purify Bgl2p to near homogeneity in one stage ().
It is important to note that the PAD-containing peptide AEGFTIFVGV (aa 80-89 “B”) contains a glutamic acid residue, which possesses a negative charge at mild alkaline pH values. Thereby this peptide at mild alkaline pH may lose its ability to fibrillate due to electrostatic repulsion between negative charges of the glutamic acid residues of the peptides. However, no significant difference was found in fibrillation of the PAD-containing peptides AEGFTIFVGV (aa 80-89 “B”) and VMANAFSYWQ (aa 187-196 “B”) at mild alkaline pH values (). Apparently the charges of amino acid residues in peptides with PADs did not impede their fibrillation significantly. On the other hand, PAD-containing peptide VDSWNVLVAG (aa 166-175 “B”) contains one residue with a negatively charged side chain (aspartic acid) and it lost its ability to fibrillate at mild alkaline conditions (). Two peptides AEGFTIFVGV (aa 80-89 “B”) and VDSWNVLVAG (aa 166-175 “B”) have each one charged residue in the second position from the N-termini of peptide. In the first case it is Glu and in the second one it is Asp. It is interesting that these peptides revealed different amyloidogenic properties in different pH values. This may be explained by the difference in the structure of the Glu residue, which differs from Asp by only a single CH2 group elongation of its side chain. This small change causes big differences in the biological uses of Asp and Glu residues. It was shown for CsgA protein that certain Asp and Gly residues functioned as gatekeeper residues which inhibited the amyloidogenic properties of these repeating units.Citation36 Certain aspartic acid residues were shown to inhibit the intrinsic aggregation tendencies of CsgA – a major subunit of the bacterial cell surface of amyloid curlin.Citation36
Alignment of predicted PADs with Bgl2p homologs (see Supplementary Information section “Search for PAD-containing peptides in Bgl2p fungi and plant homologues”) led to the hypothesis that homologs of Bgl2p from S.cerevisiae from different species (including pathogenic yeast species, e.g., C.albicans) also possessed amyloid-like properties. In a previous study we have shown that Bgl2p was not the only protein which could form amyloid in a CW of S.cerevisiae yeasts.Citation37 Our result fit well with the predictions about presence of β-aggregation prone proteins in the cell wall made in the work of Tartaglia and Caflisch.Citation38
The difference in morphology of fibrils formed by Bgl2p obtained from the CW not treated with trypsin, and the fibrils formed by Bgl2p obtained from trypsin-treated CW, was shown (). The first ones looked like “wisp” supramolecular aggregates with evident aggregation centers, and the second ones were much less ordered (“worm-like”). Both morphologies have been observed earlier for well characterized amyloid fibrils, e.g., “wisps” of glucagon fibrils and “worm-like” Aβ fibrils.Citation39,Citation40 The organization of Bgl2p fibrils may be defined by the compound (so called nucleator) susceptible to trypsin hydrolysis. Similar phenomenon is known for E. coli curli extracellular amyloid fibril polymerization which is directed by CsgB nucleator.Citation41 Adding of β-1,3-glucan (major yeast cell wall component) to Bgl2p, isolated from trypsin-treated CWs, did not reveal significant alteration of Bgl2p fibril morphology (data not shown). Electron microscopy revealed slightly more visible aggregation centers (Fig. Four I and J). These results indicated that β-1,3-glucan evidently is not strong nucleator in this case. It should be noted that trypsin did not cleave Bgl2p.Citation16 Apparently the presumptive Bgl2p nucleator (PBN) contained the peptide part with arginine and/or lysine residues which are the prerequisites of trypsin proteolysis sites.
Our studies showed a positive correlation between the predicted degree of amyloidogenicity of various Bgl2p peptide sequences and their experimentally measured ability to fibrillate. We focused on the pH-dependence of Bgl2p fibrillation. Using amyloidogenic peptides as a model for the investigation we demonstrated that pH value was the important factor, which defined the existence of Bgl2p in a soluble or an amyloid form and proved this proposition using Bgl2p isolated from CW by heating. We demonstrated also that at mild acidic and neutral pH values Bgl2p formed fibrillar structures with different morphology ( and ). We suggest that the observed fibrillation of Bgl2p was the result of PADs interaction. On the other hand the morphology of Bgl2p fibrils depended on isolation conditions and apparently on yet unidentified constituents.
Materials and Methods
Salts and low-molecular weight compounds were of analytical grade or better. Congo Red (3,3′-[(1,1’-biphenyl)-4,4’-diylbis(azo)]bis-(4-amino-1-naphthalene acid) disodium salt; Sigma, product number C6277) and ThT (4-(3,6-dimethyl-1,3-benzothiazol-3-ium-2-yl)-N,N-dimethylaniline chloride; Sigma, product number T3516) were used without further purification. TRIS was purchased from Merck (product number 648309). Qualifications and producers of the other compounds are indicated in the sections devoted to the corresponding methods. Milli-Q water was used throughout.
S. cerevisiae strain
DBY 746 (genotype MATα ura3-52 leu2-3,112 trp1-289 his3-Δ1) further referred to as “wild type” mating type α kindly provided by M.D. Ter-Avanesyan (Cardiology Research center). This strain is parental with respect to A270 strain.
A270 with the deletion in SSU21/MCD4 gene (genotype MATα ura3-52 leu2 trp1-289 his3-Δ1 ssu21) and its revertant strain bearing pHTSA plasmid with SSU21/MCD4 geneCitation42 were kindly provided by M.D. Ter-Avanesyan and G.V. Fominov (Cardiology Research center). Growing of yeasts in liquid nutritious YPD (1% yeast extract, 2% peptone, 2% glucose) medium was performed in 750 ml flasks with a growth medium volume of 200 ml in an orbital shaker (200 rpm) at 30°C.
Yeast cell wall isolation
Log-phase yeast cells (19 h of growth) were precipitated by centrifugation for 10 min at 1,650 g (OPn-8 with RU 180L rotor, Russia), washed twice with 0.05 М potassium-phosphate buffer рН 8.0 and disrupted in the shaker (Heidolph) using glass beads (0.5 mm; Sigma) on cooling. The extent of cell disruption was estimated using a light microscope (Opton). Only CW preparations containing less than 0.1% of intact cells were used in the further work. CWs were separated from the intracellular contents by centrifugation at 3,000 g for 5 min. CWs and cells formed the double-layer precipitate; CWs forming the upper layer were carefully suspended in water and separated from the cells. Then the CW preparation obtained was washed twice with water, twice with 1% saccharose, twice with 1 М NaCl, twice with 1% NaCl and once with water. According to the data obtained in our laboratory earlier, the treatments allow the complete removal of intracellular contents and cytoplasmic membrane components (Kalebina, unpublished results). The amount of CW was estimated spectrophotometrically [absorbance at 540 nm (A540) of 1 ml of 500 µg ml−1 CW suspension equaled approximately 1.0].
Yeast cell wall partial deproteinization
For Bgl2p isolation two different partial deproteinisated CW-preparations were used. One of which was subjected to trypsin and SDS treatment and the other was subjected to SDS treatment only.
Isolated CWs (A540 = 100) were suspended in 3 ml of trypsin solution (10 mg ml−1 in 0.05 М TRIS-HCl buffer, pH 7.5) (Sigma) and incubated for 2 h at 37°C. For trypsin removal, CWs were washed four times with 1 М NaCl and twice with water. To ascertain the absence of proteolytic activity in the preparation, the test with prestained casein (Reakhim) was used. In short, an aliquot of CW suspension was incubated with prestained casein solution (10 mg ml−1) for 30 min at 37°C, and then the protein was precipitated by incubation with 7% trichloroacetic acid (15 min at 4°C) and centrifugation at 11,100 g (Eppendorf Minispin). The supernatant containing proteolytic peptides conjugated with chromophore groups was separated from the precipitate and investigated colorimetrically at 400 nm. The absence of the supernatant color change was evidence of the absence of proteolytic activity in the preparation studied. The solution of prestained casein in the absence of proteases was used as a control. After complete proteinase removal, CW suspension was centrifuged at 3,000 g for 3 min, and the supernatant was discarded.
Treatment with 1% SDS (Amresco, 0227) (1 h at 37°C) was also applied. To remove SDS, the CWs obtained were additionally washed five times with 0.2 M sodium acetate buffer (pH 5.5), three times with n-butanol/water mixture 0.7:1 (vol/vol) and with water, until the smell of n-butanol had disappeared. After each ablution, the suspension was centrifuged at 2,580 g for 3 min, and the supernatant was discarded. CWs were stored at 4°C in 0.05 М TRIS-HCl buffer (pH 7.5), and sodium azide was added to the final concentration of 0.02%, if extended storage was required.
Isolation of Bgl2p glucantransferase from the CWs
Glucantransferase Bgl2p was isolated from trypsin/SDS- and SDS-treated CWs (A540 = 15) as previously described.Citation16
Protein extraction in TRIS solution
Purified CWs were incubated in 100 mM TRIS solution, pH 9.2 in ratio 1 optical unit CW (A540) for 2 µl 100 mM TRIS solution during 4.5 h at 30°C. Bgl2p extract was separated from CW by centrifugation at 13,400 rpm (Minispin) during 2 min at room temperature.
Prediction of potential amyloidogenic determinants
The Bgl2p sequence was analyzed using six computational algorithms, and the results obtained from the various algorithms were compared. In this work, the following algorithms were used: FoldAmyloid (http://bioinfo.protres.ru/fold-amyloid/oga.cgi), TANGO (http://tango.crg.es/), AGGRESCAN (http://bioinf.uab.es/aggrescan/), PASTA (http://protein.cribi.unipd.it/pasta/), WALTZ (http://waltz.switchlab.org/), as well as the secondary structure prediction server DHPRED (http://www.fz-juelich.de/nic/cbb/service/dhpred.php). The principles underlying the work of these algorithms can be found in articles devoted to these issues.Citation20-Citation28 Importantly, the average correlation between the change in aggregation propensity observed in vivo and the change in aggregation propensity predicted in silico by the algorithms FoldAmyloid and AGGRESCAN was better than for the algorithms TANGO, PASTA and WALTZ.Citation17
Thioflavin T amyloid-specific fluorescent probe assays
For fibrillation kinetic measurements of peptides with non-blocked termini or Bgl2p protein, a Fluostar Optima platereader (BMG Labtechnologies) was used with 384-well plates with optical bottoms from Nalge Nunc International (Rochester). The samples were assayed in triplicates, and each well contained 50 µl of 25 µg ml−1 peptide or 30 µg ml−1 protein sample in phosphate-citrate buffer. A final ThT concentration of 25 µM ThT (filtered) was introduced to each well. For the ThT concentration determination, a molar extinction coefficient at 412 nm of 36,000 M−1 cm−1 was applied for ThT in water. The wells were covered with Polyolefin non-sterile sealing tape, also from Nalge Nunc International, to avoid evaporation of the samples. ThT fluorescence measurements were performed as bottom/bottom measurements using an excitation wavelength of 450 nm (10 nm bandpass) and an emission wavelength of 480 nm (12 nm bandpass). The fluorescence measurements were performed at 35°C or 45°C every 400 sec. During each cycle of 400 sec, there was 1 mm orbital shaking (600 rpm) for 240 sec.
Fluorescence measurements of fibrillation of peptides with termini blocked were performed with Cary Eclipse fluorescence spectrophotometer with 5 nm excitation and emission bandpass (Varian Inc.). Excitation and emission wavelengths were the same as used for the Fluostar Optima platereader. Peptides were initially dissolved in 100% DMSO (up to concentration of peptides equal 2 mg ml−1). After that they were dissolved in phosphate-citrate buffer and final concentration of DMSO was no more than 1.25%. It should be noted that peptides with blocked termini did not fibrillate without shaking. Fibrillation of peptides with termini blocked was performed using incubator shaker Excella E24 (New Brunswick) at 35°C with 1.9 cm orbital shaking (200 rpm). The samples were assayed in triplicates in 1.5 ml tubes (each tube contained 500 µl of 25 µg ml−1 peptide in phosphate-citrate buffer). A final ThT concentration of 25 µM ThT (filtered) was introduced to each tube.
Higher pH values (more than 7.6) were excluded from the analysis due to the ThT instability under alkaline conditions.Citation43
Synthesis of PAD-containing peptides
Synthesis of peptides with non-blocked (not acetylated, not amidated) termini was performed on an n-alcoxybenzyl polymer (containing 0.5 mmol g−1 of hydroxyl groups) using standard procedures of solid phase synthesis for Fmoc\But protective groups of amino acids.Citation44 Removal of a peptide from a polymer and simultaneous unblocking of side groups of amino acid residues was conducted in a mixture of trifluoroacetic acid (TFA): thioanisole: H2O: ethanedithiol (85:5:5:5) for 2 h. The peptides were purified using reversed phase high performance liquid chromatography in a gradient of acetonitryl in 0.1% TFA (10% to 70% in 60 min) at an eluent velocity of 4 ml min−1. The eluent's absorption was registered at a wavelength of 226 nm. The HPLC experiments were performed with a System Gold chromatograph (Beckman), in Jupiter 5μ C18 300A 250 × 4.60 mm (Phenomenex) and Reprosil-Pur C18AQ 5μ 250 × 4.60 mm (Dr. Maisch, Germany) columns in the case of analytical chromatography, and in a Jupiter 10μ C18 300A 250 × 10.00 mm (Phenomenex) in the case of the preparatory one. The molecular weight of the peptides was measured with a VISION 2000 device (Bioanalysis), according to the MALDI method, and was found to correspond with the calculated one.
Synthesis of peptides with termini blocked (acetylated, amidated) was performed on a Rink-polymer using standard procedures of solid phase synthesis for Fmoc\But protective groups of amino acids.Citation44 Removal of a peptide from a polymer and simultaneous unblocking of side groups of amino acid residues was conducted in a mixture of TFA: H2O: ethanedithiol (90:5:5) for 2 h. The peptides were purified using reversed phase high performance liquid chromatography in a gradient of acetonitryl in 0.1% TFA (30 to 70% in 40 min) at an eluent velocity of 4 ml min−1. The eluent's absorption was registered at a wavelength of 226 nm. The HPLC experiments were performed with a System Gold chromatograph (Beckman), in Jupiter 5μ C4 300A 250 × 4.60 mm (Phenomenex) column in the case of analytical chromatography, and in a Jupiter 10μ C4 300A 250 × 10.00 mm (Phenomenex) in the case of the preparatory one. The molecular weight of the peptides was measured with a VISION 2000 device (Bioanalysis), according to the MALDI method, and was found to correspond with the calculated one.
In the majority of the studies the peptides with non-blocked termini were dissolved in deionized water and then buffer was added. Peptides with termini blocked were initially dissolved in dimethyl sulfoxide and then buffer was added (final concentration of dimethyl sulfoxide was no more that 1.2%). Concentrations of peptides were equilibrated to 25 µg ml−1 before experiments.
Protein and peptide concentration measurement
Protein and peptide concentrations were measured according to the technique of Scopes.Citation45 Sample absorption in the range of 190–350 nm was determined using a Cary 50 Scan UV-Visible Spectrophotometer (Varian Inc.). Bgl2p concentration was 0.03 mg ml−1.
Confocal microscopy of samples stained with ThT
Images of fibrils formed by peptides were obtained using fluorescent confocal scanning microscopes, Carl Zeiss Axiovert 200M LSM 510 META (Zeiss), or Leica TCS SP2 AOBS (Leica).
Bgl2p peptide samples placed on glass slides were incubated in conditions preventing desiccation in case of usage of phosphate-citrate buffer with pH 5.0 or 7.6. Afterwards, samples were stained with 7.5 µM of ThT. ThT was excited using an Argon laser (458 nm), and the emission signal was detected in the range of 475–525 nm.
Confocal microscopy of Bgl2p samples stained with antibody
Images of fibrils formed by glucantransferase Bgl2p were obtained using fluorescent confocal scanning microscopes Carl Zeiss Axiovert 200M LSM 510 META (Zeiss), or Leica TCS SP2 AOBS (Leica).
Bgl2p-containing samples placed at glass slides were incubated in conditions preventing desiccation. After that, samples were stained with mouse primary polyclonal antibody against Bgl2p and secondary polyclonal goat anti-mouse antibody lgG labeled with Alexa-488 fluorophore (Invitrogen).
Electron microscopy
Negative-staining EM was used. Small volumes (2 µl) of samples containing at least 60 ng of Bgl2p were adsorbed onto glow-discharged carbon-coated, Formvar-filmed 400-mesh copper grids and immediately dried down. 2% uranyl acetate staining solution was then absorbed for 2 min. Grids were allowed to dry in a light-protected environment and were viewed in a JEM-100B (JEOL) electron microscope at the accelerating potential of 80 kV.
Electrophoresis, western blot analysis
Electrophoresis was performed according to LaemmliCitation46 in 10% resolving polyacrylamide gels. Proteins were stained with Coomassie G-250 or immunologically detected by Western Blot analysis. Prestained Protein Molecular Weight Marker (Fermentas, Canada) was used. Bgl2p antiserum was raised in male BALB/c mice (SPF status) using SDS PAGE-purified protein (40 μg per mouse).Citation47
Other methods
S. cerevisiae ssu21/mcd4 mutant growth medium ultrafiltration was performed using MidGee Cross Flow hollow fiber cartridges (GE Healthcare) with hold-up volume of 0.5 ml and cut-off limit of 100 kDa and using Millipore filters with cut-off limit of 10 kDa.
| Abbreviations: | ||
| “B” | = | blocked peptides’ termini (acetylated, amidated) |
| Bgl2p | = | glucantransferase Bgl2p |
| Bgl2pGM | = | glucantransferase Bgl2p from growth media |
| CR | = | Congo Red |
| CW | = | cell wall |
| "NB" | = | non-blocked peptides’ termini (not acetylated, not amidated) |
| PAD | = | potential amyloidogenic determinant |
| TFA | = | trifluoroacetic acid |
| ThT | = | thioflavin T |
Additional material
Download Zip (617.7 KB)Acknowledgments
This work was funded by the Carl Zeiss Program for Support of Research Work of Young Investigators from Higher Education Institutions of Russia (Grant no. MSU 16/11CZ), the Drug Research Academy (Faculty of Pharmaceutical Sciences, University of Copenhagen), the Russian Foundation for Basic Research (Grant no. 09-04-00768-a, 10-04-01821a, 11-04-763a, 12-04-31966-mol_a), the programs “Molecular and Cellular Biology” (01200959110 and 01200957492) and “Fundamental Sciences to Medicine,” Howard Hughes Medical Institute (55005607), the Federal Agency for Science and Innovation (grant no. 02.740.11.0295 and 16.740.11.0478) and by the German organization DAAD (German Academic Exchange Service) in the frame of a Scientific Cooperation Agreement between Moscow and Rostock Universities. The authors would like to acknowledge associate professor Osman Mirza (Faculty of Health and Medical Sciences, University of Copenhagen) for providing access to facilities useful for growing yeast, professor Per Amstrup Pedersen (Department of Biology, University of Copenhagen) for the BMA64-1B strain, Stefano Colombo (Faculty of Health and Medical Sciences, University of Copenhagen) for discussion of the results obtained, Dr S.N. Egorova and Dr M.N. Zhmak (Institute of Bioorganic Chemistry, Moscow, Russia) for synthesis of the peptides, and Dr Annette E. Langkilde (Faculty of Health and Medical Sciences, University of Copenhagen) for collecting the X-ray fiber diffraction data. We are grateful to Tatyana A. Sabirzyanova for the discussion of our results and to Aleksandra Zhukova for the assistance in preparing of manuscript.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Notes
† Current affiliation: Laboratory of Biochemistry and Genetics; National Institute of Diabetes and Digestive and Kidney Disease; National Institutes of Health; Bethesda, MD USA
References
- Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem 2006; 75:333 - 66; http://dx.doi.org/10.1146/annurev.biochem.75.101304.123901; PMID: 16756495
- Westermark P, Benson MD, Buxbaum JN, Cohen AS, Frangione B, Ikeda S, et al. A primer of amyloid nomenclature. Amyloid 2007; 14:179 - 83; http://dx.doi.org/10.1080/13506120701460923; PMID: 17701465
- Wösten HA, de Vocht ML. Hydrophobins, the fungal coat unravelled. Biochim Biophys Acta 2000; 1469:79 - 86; http://dx.doi.org/10.1016/S0304-4157(00)00002-2; PMID: 10998570
- Chapman MR, Robinson LS, Pinkner JS, Roth R, Heuser J, Hammar M, et al. Role of Escherichia coli curli operons in directing amyloid fiber formation. Science 2002; 295:851 - 5; http://dx.doi.org/10.1126/science.1067484; PMID: 11823641
- Shewmaker F, McGlinchey RP, Wickner RB. Structural insights into functional and pathological amyloid. J Biol Chem 2011; 286:16533 - 40; http://dx.doi.org/10.1074/jbc.R111.227108; PMID: 21454545
- Fowler DM, Koulov AV, Alory-Jost C, Marks MS, Balch WE, Kelly JW. Functional amyloid formation within mammalian tissue. PLoS Biol 2006; 4:e6; http://dx.doi.org/10.1371/journal.pbio.0040006; PMID: 16300414
- Larsen P, Nielsen JL, Dueholm MS, Wetzel R, Otzen D, Nielsen PH. Amyloid adhesins are abundant in natural biofilms. Environ Microbiol 2007; 9:3077 - 90; http://dx.doi.org/10.1111/j.1462-2920.2007.01418.x; PMID: 17991035
- Otzen D, Nielsen PH. We find them here, we find them there: functional bacterial amyloid. Cell Mol Life Sci 2008; 65:910 - 27; http://dx.doi.org/10.1007/s00018-007-7404-4; PMID: 18034321
- Gebbink MF, Claessen D, Bouma B, Dijkhuizen L, Wösten HA. Amyloids--a functional coat for microorganisms. Nat Rev Microbiol 2005; 3:333 - 41; http://dx.doi.org/10.1038/nrmicro1127; PMID: 15806095
- Teertstra WR, van der Velden GJ, de Jong JF, Kruijtzer JA, Liskamp RM, Kroon-Batenburg LM, et al. The filament-specific Rep1-1 repellent of the phytopathogen Ustilago maydis forms functional surface-active amyloid-like fibrils. J Biol Chem 2009; 284:9153 - 9; http://dx.doi.org/10.1074/jbc.M900095200; PMID: 19164282
- Otoo HN, Lee KG, Qiu W, Lipke PN. Candida albicans Als adhesins have conserved amyloid-forming sequences. Eukaryot Cell 2008; 7:776 - 82; http://dx.doi.org/10.1128/EC.00309-07; PMID: 18083824
- Teertstra WR, Krijgsheld P, Wösten HA. Absence of repellents in Ustilago maydis induces genes encoding small secreted proteins. Antonie Van Leeuwenhoek 2011; 100:219 - 29; http://dx.doi.org/10.1007/s10482-011-9581-2; PMID: 21626092
- Lundmark K, Westermark GT, Olsén A, Westermark P. Protein fibrils in nature can enhance amyloid protein A amyloidosis in mice: Cross-seeding as a disease mechanism. Proc Natl Acad Sci U S A 2005; 102:6098 - 102; http://dx.doi.org/10.1073/pnas.0501814102; PMID: 15829582
- Klebl F, Tanner W. Molecular cloning of a cell wall exo-beta-1,3-glucanase from Saccharomyces cerevisiae.. J Bacteriol 1989; 171:6259 - 64; PMID: 2509432
- Mrša V, Klebl F, Tanner W. Purification and characterization of the Saccharomyces cerevisiae BGL2 gene product, a cell wall endo-beta-1,3-glucanase. J Bacteriol 1993; 175:2102 - 6; PMID: 8458852
- Kalebina TS, Plotnikova TA, Gorkovskii AA, Selyakh IO, Galzitskaya OV, Bezsonov EE, et al. Amyloid-like properties of Saccharomyces cerevisiae cell wall glucantransferase Bgl2p: prediction and experimental evidences. Prion 2008; 2:91 - 6; http://dx.doi.org/10.4161/pri.2.2.6645; PMID: 19098439
- Belli M, Ramazzotti M, Chiti F. Prediction of amyloid aggregation in vivo.. EMBO Rep 2011; 12:657 - 63; http://dx.doi.org/10.1038/embor.2011.116; PMID: 21681200
- López De La Paz M, Goldie K, Zurdo J, Lacroix E, Dobson CM, Hoenger A, et al. De novo designed peptide-based amyloid fibrils. Proc Natl Acad Sci U S A 2002; 99:16052 - 7; http://dx.doi.org/10.1073/pnas.252340199; PMID: 12456886
- Ohhashi Y, Hasegawa K, Naiki H, Goto Y. Optimum amyloid fibril formation of a peptide fragment suggests the amyloidogenic preference of β2-microglobulin under physiological conditions. J Biol Chem 2004; 279:10814 - 21; http://dx.doi.org/10.1074/jbc.M310334200; PMID: 14699107
- Galzitskaya OV, Garbuzynskiy SO, Lobanov MY. Prediction of amyloidogenic and disordered regions in protein chains. PLoS Comput Biol 2006; 2:e177; http://dx.doi.org/10.1371/journal.pcbi.0020177; PMID: 17196033
- Garbuzynskiy SO, Lobanov MY, Galzitskaya OV. FoldAmyloid: a method of prediction of amyloidogenic regions from protein sequence. Bioinformatics 2010; 26:326 - 32; http://dx.doi.org/10.1093/bioinformatics/btp691; PMID: 20019059
- Galzitskaya OV, Garbuzynskiy SO, Lobanov MY. Is it possible to predict amyloidogenic regions from sequence alone?. J Bioinform Comput Biol 2006; 4:373 - 88; http://dx.doi.org/10.1142/S0219720006002004; PMID: 16819789
- Fernandez-Escamilla AM, Rousseau F, Schymkowitz J, Serrano L. Prediction of sequence-dependent and mutational effects on the aggregation of peptides and proteins. Nat Biotechnol 2004; 22:1302 - 6; http://dx.doi.org/10.1038/nbt1012; PMID: 15361882
- Sánchez de Groot N, Pallarés I, Avilés FX, Vendrell J, Ventura S. Prediction of “hot spots” of aggregation in disease-linked polypeptides. BMC Struct Biol 2005; 5:18; http://dx.doi.org/10.1186/1472-6807-5-18; PMID: 16197548
- Conchillo-Solé O, de Groot NS, Avilés FX, Vendrell J, Daura X, Ventura S. AGGRESCAN: a server for the prediction and evaluation of “hot spots” of aggregation in polypeptides. BMC Bioinformatics 2007; 8:65; http://dx.doi.org/10.1186/1471-2105-8-65; PMID: 17324296
- Trovato A, Chiti F, Maritan A, Seno F. Insight into the structure of amyloid fibrils from the analysis of globular proteins. PLoS Comput Biol 2006; 2:e170; http://dx.doi.org/10.1371/journal.pcbi.0020170; PMID: 17173479
- Maurer-Stroh S, Debulpaep M, Kuemmerer N, Lopez de la Paz M, Martins IC, Reumers J, et al. Exploring the sequence determinants of amyloid structure using position-specific scoring matrices. Nat Methods 2010; 7:237 - 42; http://dx.doi.org/10.1038/nmeth.1432; PMID: 20154676
- Zimmermann O, Hansmann UH. Support vector machines for prediction of dihedral angle regions. Bioinformatics 2006; 22:3009 - 15; http://dx.doi.org/10.1093/bioinformatics/btl489; PMID: 17005536
- Groenning M. Binding mode of Thioflavin T and other molecular probes in the context of amyloid fibrils-current status. J Chem Biol 2010; 3:1 - 18; http://dx.doi.org/10.1007/s12154-009-0027-5; PMID: 19693614
- Frid P, Anisimov SV, Popovic N. Congo red and protein aggregation in neurodegenerative diseases. Brain Res Rev 2007; 53:135 - 60; http://dx.doi.org/10.1016/j.brainresrev.2006.08.001; PMID: 16959325
- Tzotzos S, Doig AJ. Amyloidogenic sequences in native protein structures. Protein Sci 2010; 19:327 - 48; http://dx.doi.org/10.1002/pro.314; PMID: 20027621
- Inouye H, Kirschner DA. X-Ray fiber and powder diffraction of PrP prion peptides. Adv Protein Chem 2006; 73:181 - 215; http://dx.doi.org/10.1016/S0065-3233(06)73006-6; PMID: 17190614
- Sawaya MR, Sambashivan S, Nelson R, Ivanova MI, Sievers SA, Apostol MI, et al. Atomic structures of amyloid cross-beta spines reveal varied steric zippers. Nature 2007; 447:453 - 7; http://dx.doi.org/10.1038/nature05695; PMID: 17468747
- Sánchez de Groot N, Pallarés I, Avilés FX, Vendrell J, Ventura S. Prediction of “hot spots” of aggregation in disease-linked polypeptides. BMC Struct Biol 2005; 5:18; http://dx.doi.org/10.1186/1472-6807-5-18; PMID: 16197548
- Kalebina TS, Egorov SN, Arbatskii NP, Bezsonov EE, Gorkovskii AA, Kulaev IS. The role of high-molecular-weight polyphosphates in activation of glucan transferase Bgl2p from Saccharomyces cerevisiae cell wall. Dokl Biochem Biophys 2008; 420:142 - 5; http://dx.doi.org/10.1134/S1607672908030125; PMID: 18680912
- Wang X, Zhou Y, Ren JJ, Hammer ND, Chapman MR. Gatekeeper residues in the major curlin subunit modulate bacterial amyloid fiber biogenesis. Proc Natl Acad Sci U S A 2010; 107:163 - 8; http://dx.doi.org/10.1073/pnas.0908714107; PMID: 19966296
- Gorkovskii AA, Bezsonov EE, Plotnikova TA, Kalebina TS, Kulaev IS. Revealing of Saccharomyces cerevisiae yeast cell wall proteins capable of binding thioflavin T, a fluorescent dye specifically interacting with amyloid fibrils. Biochemistry (Mosc) 2009; 74:1219 - 24; http://dx.doi.org/10.1134/S0006297909110066; PMID: 19916936
- Tartaglia GG, Caflisch A. Computational analysis of the S. cerevisiae proteome reveals the function and cellular localization of the least and most amyloidogenic proteins. Proteins 2007; 68:273 - 8; http://dx.doi.org/10.1002/prot.21427; PMID: 17407164
- Andersen CB, Yagi H, Manno M, Martorana V, Ban T, Christiansen G, et al. Branching in amyloid fibril growth. Biophys J 2009; 96:1529 - 36; http://dx.doi.org/10.1016/j.bpj.2008.11.024; PMID: 19217869
- Harper JD, Lieber CM, Lansbury PT Jr.. Atomic force microscopic imaging of seeded fibril formation and fibril branching by the Alzheimer’s disease amyloid-beta protein. Chem Biol 1997; 4:951 - 9; http://dx.doi.org/10.1016/S1074-5521(97)90303-3; PMID: 9427660
- Hammer ND, Schmidt JC, Chapman MR. The curli nucleator protein, CsgB, contains an amyloidogenic domain that directs CsgA polymerization. Proc Natl Acad Sci U S A 2007; 104:12494 - 9; http://dx.doi.org/10.1073/pnas.0703310104; PMID: 17636121
- Fominov GV, Ter-Avanesian MD. [Caffeine sensitivity of the yeast Saccharomyces cerevisiae MCD4 mutant is related to alteration of calcium homeostasis and degradation of misfolded proteins]. Mol Biol (Mosk) 2005; 39:464 - 76; http://dx.doi.org/10.1007/s11008-005-0056-2; PMID: 15981576
- Foderà V, Librizzi F, Groenning M, van de Weert M, Leone M. Secondary nucleation and accessible surface in insulin amyloid fibril formation. J Phys Chem B 2008; 112:3853 - 8; http://dx.doi.org/10.1021/jp710131u; PMID: 18311965
- Wellings DA, Atherton E. Standard Fmoc protocols. Methods Enzymol 1997; 289:44 - 67; http://dx.doi.org/10.1016/S0076-6879(97)89043-X; PMID: 9353717
- Scopes RK. Measurement of protein by spectrophotometry at 205 nm. Anal Biochem 1974; 59:277 - 82; http://dx.doi.org/10.1016/0003-2697(74)90034-7; PMID: 4407487
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970; 227:680 - 5; http://dx.doi.org/10.1038/227680a0; PMID: 5432063
- Morenkov OS, Mantsygin IuA, Sergeev VA, Sobko IuA, Morenkova MA, Panchenko OA. [The isolation and characteristics of monoclonal antibodies to the glycoprotein GII of Aujeszky’s disease virus and their use for the epitopic mapping of GII]. Vopr Virusol 1994; 39:174 - 7; PMID: 7527989