Abstract
Myocilin is a protein with a molecular weight near 50 kDa. It is expressed in almost all organs and tissues.Citation1 We showed that the peptide DQL ETQ TRE LET AYS NLL RD corresponding to N-terminal Leucine zipper motif (LZM) of the protein is able to form amyloid-like fibrils. The possible role of this motif in myocilin aggregation is discussed.
Keywords: :
Introduction
Myocilin function currently remains unknown.Citation2 There is a hereditary form of glaucoma caused by mutations in the myocilin gene, leading to a change in its primary structure.Citation3 The mutant forms of myocilin show increased tendency for aggregation. In recent studies it was shown that mutated myocilin can form amyloid-like fibrils in vitro an in cell cultures. Possible amyloidogenic motif is located at 228–244 position of the C-terminal (227–504) domain of the protein.Citation4 Analysis of the myocilin primary structure shows that the N-terminal (myosin-like) domain of the protein contains a potential mitochondrial localization signal and LZM.Citation5 LZMs are able to participate in protein-protein interactions and in homooligomerization. It can be considered as another possible amyloidogenic determinant in this protein. In our work we studied the homooligomerisation ability of an artificial peptide, containing the LZM and corresponding to the primary structure of 129–148 fragment of human myocilin.
Results
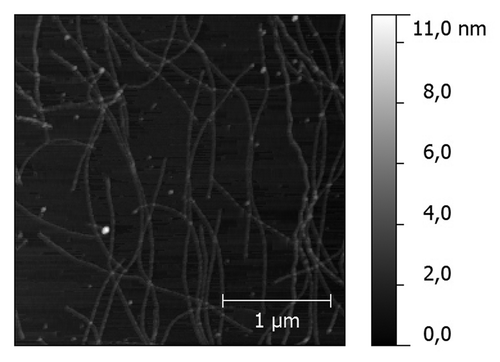
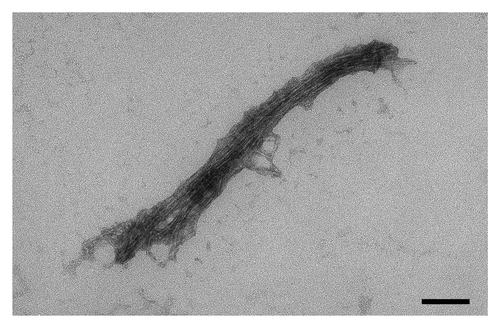
Incubation of the peptide DQL ETQ TRE LET AYS NLL RD for two hours in PBS (pH 7.0) at 37ºC led to a noticeable opalescence of the peptide solution. Peptide solution was then applied to freshly cleaved mica and examined by atomic force microscopy. The results of atomic force microscopy showed a presence of a net of fibrillar aggregates of various sizes (). Transmission electron microscopy with standard negative staining techniques revealed that the aggregates are formed by bundles of fine fibrils ().
Figure 2. Transmission electron microscopy of the aggregates shown in . Black box in the bottom-right corner corresponds to 100 nm.
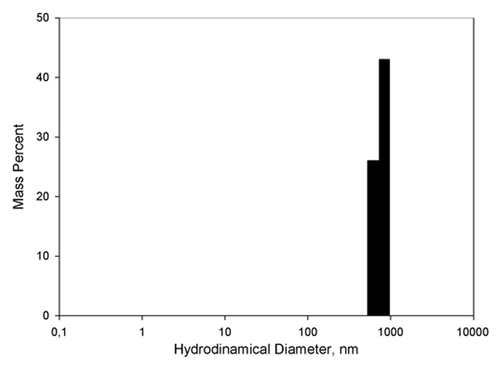
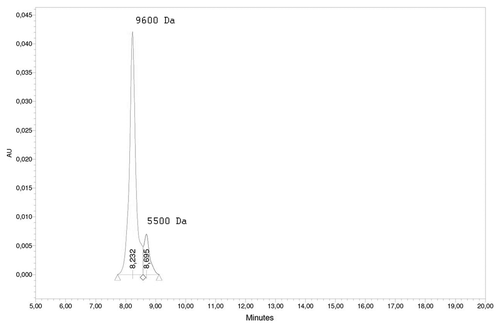
Quasielastic light scattering experiments showed that the peptide solution contains small objects with hydrodynamic diameter around 700–1000 nm, accounting for approximately 70% of the total mass of the solute (). Measurements of the concentration of the peptide in the supernatant after centrifugation of the peptide solution at 12000 g have also shown that near 70% of the peptide have formed aggregates large enough to precipitate by such centrifugation. We also performed an analytical HPLC for characterization of the non-precipitated peptide () and observed two peaks, likely corresponding to di- (2.3*MWmonomer) and tetramers (4*MWmonomer) of the peptide.
Figure 4. Analytical HPLC of the supernatant after centrifugation of the peptide solution at 12000 g for one minute. Peak 8.232 min corresponds to MW of 9600 Da, peak 8.695 corresponds to 5500 Da. Molecular weight of the peptide monomer is 2395 Da.
The precipitate of the peptide aggregates was further studied with the use of the amyloid-specific dyes ThT and Congo Red (CR). Confocal imaging of the precipitated peptide aggregates stained with ThT () clearly show areas of high ThT fluorescence. We also used optical microscopy to study CR-stained aggregates under polarized light. Green birefringence was observed on the edges and on the surface of the spherulite-like aggregate from the peptide precipitate (). These measurements provide a strong evidence for an amyloid nature of the aggregation of DQL ETQ TRE LET AYS NLL RD peptide.
Figure 5. Fluorescence spectrum of the peptide fibrils complexes with Thioflavine T (excitation 440 nm) and confocal microscopy of the peptide precipitate stained with Thioflavine T. Left panel: fluorescence spectrum of 10 mkg/ml peptide solution with 2 mM ThT in 50 mM phosphate buffer pH 6.0. Right panel (field 50 × 50 μm): the confocal image of the aggregate with ThT.
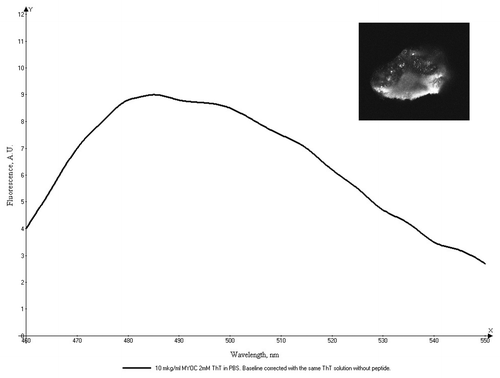
Figure 6. Optical microscopy of aggregates of the Leu-Zipper containing peptide stained with Congo Red under the cross-polarized light. Green birefringence present on the surface and edges of the aggregate.
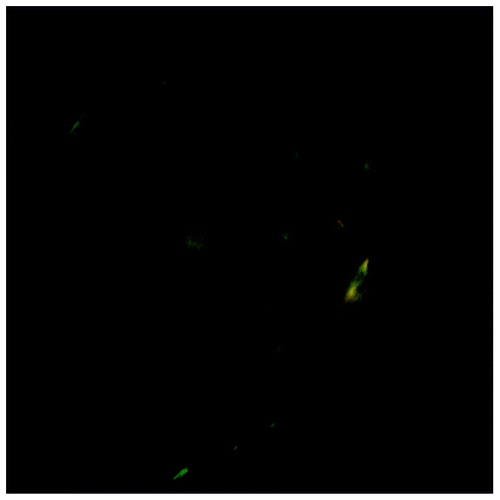
Our results indicate that the synthetic peptide corresponding to LZM of myocilin not only exhibit tendency to oligomerisation and aggregation, but also appears to form β-sheet amyloid-like structures.
Discussion
The ability of DQL ETQ TRE LET AYS NLL RD peptide to form amyloid-like fibrils suggests that, in addition to OLF-like C-domain,Citation4 LZM located in the N-terminal domain is a possible amyloidogenic site in myocilin. The classical LZM-containing peptides usually form α-helix structure, however, depending on the properties of the amino acids present between leucines it can be stable at β-conformation.Citation6 According to the existing three-dimensional model of myocilin,Citation7 we can hypothesize that LZM is located in a close proximity with the C-terminal domain of the wild-type protein and could take part in intramolecular interaction. Due to increased structural instability of the mutant protein,Citation8 LZM could become opened for intermolecular interactions and take part in the aggregation process. We can suppose that the substances which are able to specifically block such interactions may be used as new tools of molecular medicine to fight the hereditary forms of primary open-angle glaucoma.
Conclusions
We present evidence that synthetic peptide DQL ETQ TRE LET AYS NLL RD corresponding to the LZM of myocilin can form amyloid-like fibrils, providing a possible amyloidogenic site in this protein.
Materials and Methods
Materials
DQL ETQ TRE LET AYS NLL RD peptide was synthesized in the NPF “Verta” with the purity of more than 80%. Thioflavin T and reagents for electron microscopy were purchased from Sigma, USA.
Measurements of the peptide concentration
Peptide concentrations were measured spectrophotometrically by absorption at 280 nm using the theoretically predicted molar extinction coefficient of 1490 M−1 cm−1.Citation9
Incubation of the peptide
Peptide was incubated at a concentration of 0.5 mM in the phosphate buffered saline (PBS) at 37∫C on an orbital shaker Eppendorf, rotation intensity 600 rpm, incubation time 2 h. Test tubes by “Eppendorf” (0.5 ml) were used.
Atomic force microscopy
Atomic force microscopy was performed on the Bio-Solver Pro microscope with Smena-B measuring head in tapping mode. Cantilevers NSG03 (NT-MDT) were used. The samples were loaded onto freshly cleaved mica, incubated for 1 min and then washed with plenty of MilliQ deionized water. Image processing was performed using Gwyddion software.Citation10
Electron microscopy
Electron microscopy was performed on a JEOL JEM 1,000 microscope using the standard negative staining technique with phosphotungstic acid salts.
Quasi-elastic light scattering (QLS)
QLS measurements were performed using QLS-spectrometer LCS-03 (Intox). Spectra analysis was performed with the spectrometer software using “rod-like” model of the scattering objects. For measuring the 0.5 mM peptide solution was diluted 30 times with PBS to avoid double scattering.
Analytical HPLC
Peptide solution (0.5 mM in PBS) was centrifuged at 12,000 g for one minute. The supernatant was analyzed by analytical HPLC using Waters HPLC system (2489 UV/Visible detector, 1525 HPLC pump and 2707 autosampler) with Bio-Sil SEC 125–5 Column (Bio-Rad). Calibration was performed with appropriate Bio-Rad standards and set of peptides with molecular masses from 200 to 2,000 Da. Molecular weight analysis was performed with Waters software supplied with the HPLC system.
Fluorescence measurements
Measurement of fluorescence was performed on the Hitachi F-4010 spectrofluorometer with excitation on 440 nm. The 10 mkl of fibrils solution was added to 2 mM ThT in 50 mM phosphate pH 6.0. The final peptide concentration was 10 mkg/ml. The spectrum baseline was corrected with the spectrum of the same ThT solution without the peptide.
Confocal microscopy
For confocal microscopy studies the ThT stock solution was added to homogeneous 0.5 mM peptide solution to the final concentration of 10 mM. After five minutes of incubation the mixture was centrifuged at 12,000 g for one minute and then the precipitate was analyzed by a laser scanning confocal microscope (LSM; Carl Zeiss LSM 510 META). A drop of sample was put on the slide under the coverslip and was studied with an immersion objective PlanApochromat 63× = 1.4 Oil (Carl Zeiss), using a 458 nm laser for excitation of the ThT fluorescence and a 475–515 nm optical filter for fluorescence detection. Images were obtained with the help of LSM 510 software supplied with the LSM.
Congo Red (CR) staining
For the optical microscopy studies the CR stock solution was added to the homogeneous 0.5 mM peptide solution to a final CR concentration of 20 mM. After five minutes of incubation the mixture was centrifuged at 12,000 g for one minute and then the precipitate was analyzed by optical microscope. A drop of sample was put on a slide under the coverslip and placed between two polarizing plates with perpendicular directions of polarization. Analysis was performed using Leica DM-1000 optical microscope with Hi Plan 10×/0.25 objective.
| Abbreviations: | ||
| CR | = | Congo Red |
| HPLC | = | high performance liquid chromatography |
| LZM | = | leucine zipper motif |
| QLS | = | quasielastic light scattering |
| ThT | = | thioflavine T |
Acknowledgments
This study is supported by RFBR grants Nº09-04-00788-a, 07-04-01454-a, 07-04-00476-a.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Nguyen TD, Chen P, Huang WD, Chen H, Johnson D, Polansky JR. Gene structure and properties of TIGR, an olfactomedin-related glycoprotein cloned from glucocorticoid-induced trabecular meshwork cells. J Biol Chem 1998; 273:6341 - 50; http://dx.doi.org/10.1074/jbc.273.11.6341; PMID: 9497363
- Aroca-Aguilar J-D, Sánchez-Sánchez F, Martínez-Redondo F, Coca-Prados M, Escribano J. Heterozygous expression of myocilin glaucoma mutants increases secretion of the mutant forms and reduces extracellular processed myocilin. Mol Vis 2008; 14:2097 - 108; PMID: 19023451
- Wentz-Hunter K, Ueda J, Shimizu N, Yue BY. Myocilin is associated with mitochondria in human trabecular meshwork cells. J Cell Physiol 2002; 190:46 - 53; http://dx.doi.org/10.1002/jcp.10032; PMID: 11807810
- Orwig SD, Perry CW, Kim LY, Turnage KC, Zhang R, Vollrath D, et al. Amyloid fibril formation by the glaucoma-associated olfactomedin domain of myocilin. J Mol Biol 2012; 421:242 - 55; http://dx.doi.org/10.1016/j.jmb.2011.12.016; PMID: 22197377
- Wentz-Hunter K, Ueda J, Yue BY. Protein interactions with myocilin. Invest Ophthalmol Vis Sci 2002; 43:176 - 82; PMID: 11773029
- Ciani B, Hutchinson EG, Sessions RB, Woolfson DN. A designed system for assessing how sequence affects alpha to beta conformational transitions in proteins. J Biol Chem 2002; 277:10150 - 5; http://dx.doi.org/10.1074/jbc.M107663200; PMID: 11751929
- Kanagavalli J, Krishnadas SR, Pandaranayaka E, Krishnaswamy S, Sundaresan P. Evaluation and understanding of myocilin mutations in Indian primary open angle glaucoma patients. Mol Vis 2003; 9:606 - 14; PMID: 14627955
- Burns JN, Turnage KC, Walker CA, Lieberman RL. The stability of myocilin olfactomedin domain variants provides new insight into glaucoma as a protein misfolding disorder. Biochemistry 2011; 50:5824 - 33; http://dx.doi.org/10.1021/bi200231x; PMID: 21612213
- http://web.expasy.org/protparam/
- http://gwyddion.net/