Abstract
Murine prion protein deleted for residues 105–125 is intrinsically neurotoxic and mediates a TSE-like phenotype in transgenic mice. Equivalent and overlapping deletions were expressed in E.coli, purified and analyzed. Among mutants spanning the region 95–135, a construct lacking solely residues 105–125 had distinct properties when compared with the full-length prion protein 23–231 or other deletions. This distinction was also apparent followed expression in eukaryotic cells. Unlike the full-length protein, all deletion mutants failed to bind to synthetic membranes in vitro. These data suggest a novel structure for the 105–125 deleted variant that may relate to its biological properties.
Introduction
The mammalian prion protein PrPC is a GPI anchored membrane protein associated with the outer leaflet of the cell cytosolic membrane.Citation1 It is central to the spongiform encephalopathies which are protein misfolding diseases of inherited, sporadic, iatrogenic or zoonotic origin.Citation2,Citation3 Disease is marked by the conversion of PrPC to a misfolded isoform, PrPSc, and is typically associated with amyloid plaque formation, neurodegeneration and brain spongiosis. However, cases in which high levels of PrPSc occur without clinical diseaseCitation4 or without the hallmark protease resistant PrP27–30 core of PrPSc have been also describedCitation5,Citation6 leading to the suggestion that the neurotoxic species is an intermediate isoform on the folding pathway between PrPC and PrPSc.Citation7 Toxicity experiments using artificially assembled PrPSc fibrils in vitro have suggested that toxicity resides in small oligomeric species,Citation8 an off-pathway intermediate to the formation of fibrillar PrPSc,Citation9 plausibly explaining the uncoupling of infectivity from PrPSc titer and pathogenicity in some cases.Citation10
At the molecular level, a hydrophobic span of residues (HP) within the region 105–125 of the intrinsically disordered N-terminus of PrPC has been shown to mediate pathogenicity in vitro and in vivo and a peptide comprising the PrPC sequence 106–126 exhibits conditional toxicity when added to neuronal cell lines expressing WT PrPC.Citation11 Further, transgenic mice heterozygous for a PrPC gene encoding a deletion of the HP sequence (PrP Δ105–125) display a neonatally lethal phenotype that is rescued by the overexpression of WT PrPC.Citation12,Citation13 WT PrPC and PrP Δ105–125 have comparative expression levels in cell culture and both localize to the outer leaflet of the cytosolic membrane suggesting that the toxicity of PrP Δ105–125 is not due to instability within the cell.Citation14 Instead, a mechanism has been proposed in which the deleterious effects of PrP Δ105–125 are rescued by PrPC WT through competition for a common membrane surface receptor that may mediate inward pro/anti-apoptotic signaling dependant on the PrPC isoform that is bound. Thus, PrP Δ105–125 could subvert normal function to mediate pathogenicity.Citation15
An alternative model of PrP Δ105–125 toxicity has also been described in which PrP Δ105–125 facilitates spontaneous inward ionic currents that are absent in cell lines expressing WT PrPC alone and are suppressed by its co-expression.Citation16 Both models suggest that PrP Δ105–125 adopts a different conformation to that of WT PrPC and this is supported in part by available biochemical and biophysical measurement.Citation17 To explore further the conformational properties of PrP Δ105–125 compared with PrPC WT an extended set of deletion mutants in the hydrophobic region of murine prion protein were constructed and the resulting proteins subjected to comparative biophysical analyses following expression and purification.
Results
To probe the role of residues 105–125 in PrPC conformation, a series of overlapping deletions was constructed in the context of mouse PrPC (NM_011170) () and the resulting sequences expressed as His tagged proteins in E.coli. Each protein could be purified to homogeneity by a combination of differential solubility and metal chelate chromatography (). Following refolding all PrPC variants were found to be soluble and suitable for a low resolution structural probe analysis using the environmentally sensitive fluorescent dye 1-anilinonaphthalene 8-sulfonate (ANS) to assess the solvent accessibility of hydrophobic residues.Citation18-Citation20 As PrPC is a Cu2+ binding proteinCitation21-Citation23 ANS measurements were performed in the presence and absence of 50μM CuSO4. The observed ANS fluorescence was found to be inversely proportional to the extent of the deletion introduced with the notable exception of the Δ105–125 construct which displayed a fluorescence value similar to that of WT 23–231 (). The same relative ANS fluorescence was observed when all samples were measured in the presence of Cu2+ although the levels were lower overall, consistent with Cu2+ bound PrP molecules adopting a more compact fold and allowing less access to hydrophobic residues.Citation24,Citation25
Figure 1. Design and expression of prion deletion mutants. (A) Schematic representation of murine PrP constructs. The N-terminal basic region is between amino acids 23 and 31. The five octarepeats are between amino acids 51 and 90. The charged cluster upstream of the hydrophobic region is indicated by the box between amino acids 95 and 105. The neurotoxic hydrophobic region is boxed between amino acids 105 and 125. Beta sheets 1 and 2 are labeled S1 and S2, and helices 1, 2 and 3 as H1, H2 and H3 respectively. Balls on sticks represent the glycosylation sites at Asn 180 and Asn 196. The disulphide bridge linking helicies 2 and 3 at Cys 179 and Cys 214 is shown as S-S. (B) SDS-PAGE analysis of all purified prion proteins. Lane 1, PrP 90–231; lane 2, PrP 23–231; lane 3, PrP Δ95–105; lane 4, PrP Δ95–125; lane 5, PrP Δ95–135; lane 6, PrP Δ105–125; lane 7, PrP Δ105–135; lane 8, 5µg of BSA. Numbers to the left of 1B are molecular mass markers and are in kilodaltons (kDa).
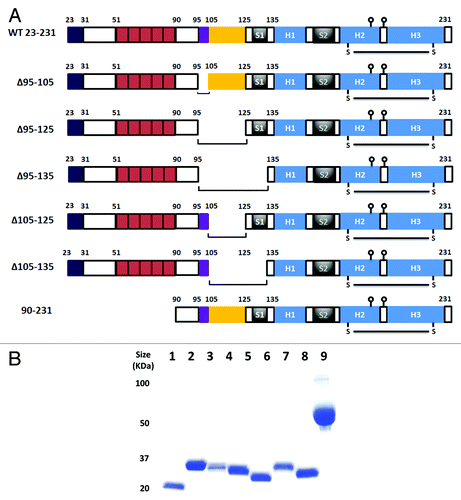
Figure 2. ANS fluorescence. (A) Each prion construct was refolded and assessed for ANS binding in the presence and absence of Cu2+. The concentration of proteins was 35µM, the ANS concentration 600µM and the Cu2+ concentration 50µM. All readings were normalized to control measurement of buffer with ANS alone. Measurements were performed in triplicate and error bars indicate the standard error mean. (B) The effect of pH on ANS fluorescence for WT 23–231 (circle), Δ105–125 (square) and 90–231 (diamond). The filled symbols/solid lines are ANS data in the absence of Cu2+ while the open symbols/dashed lines are ANS data in the presence of Cu2+. The assay conditions were as above with the exception that the pH was independently buffered in each case. Experiments were performed in triplicate and error bars indicate the standard error mean.
The novel hydrophobic solvent accessibility of PrPC Δ105–125 in relation to the other deletion mutants was explored further. As copper loading of PrPC leads to its endocytosis and trafficking to early endosomesCitation22,Citation23,Citation26 where the pH is more acidic,Citation27 the instability of PrP Δ105–125 in acidic pH might contribute to its observed toxicity. Thus, PrP Δ105–125, WT 23–231 and PrP 90–231 (representing a negative control for Cu2+ binding as it lacks the octarepeats) were assayed for ANS binding in a pH range from 3–7 in the presence and absence of Cu2+ as before. Lowering the pH led to an increase in solvent exposed hydrophobic residues for all constructs tested but Δ105–125 displayed the highest overall ANS fluorescence across the pH range (). The presence of Cu2+ ions lowered ANS florescence for all constructs to a value pH 5 as before (compare ) but below pH 5 ANS fluorescence markedly increased, suggesting general unfolding.
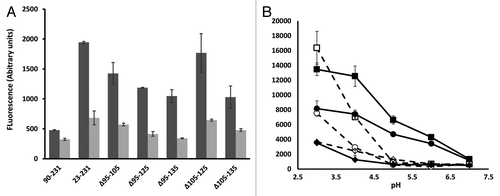
To assess whether the increased ANS fluorescence of PrP Δ105–125 compared with WT 23–231 was also associated with a change in overall structure as reflected by epitope accessibility to a number of monoclonal antibodies (MAbs), each refolded protein was assessed by comparative ELISA with the anti-prion MAbs 6D11,Citation28 7H6,Citation29 6H4,Citation30 8H4 and 9H7,Citation29 none of which have published epitopes within the deleted region (). All MAbs bound similarly to each protein with the exception of 7H6 which failed to bind appreciably to Δ105–125 (). The epitope for 7H6 has been reported to lie within residues 130–140Citation29 which, of all the MAbs tested, is the epitope closest to the region deleted in Δ105–125 so the possibility that binding to the epitope is affected by its proximity to the deleted sequence cannot be excluded. Overall however, there was no evidence for a substantial change in accessibility to the polypeptide backbone of Δ105–125 as measured by antibody binding. Circular dichroism (CD) spectroscopy was used to probe the secondary structure of each purified deletion mutant after normalization of protein concentration and the obtained spectra were deconvoluted using the K2D3 algorithm.Citation31 The deletion mutants generally gave weaker CD spectra than WT 23–321 and PrP 90–231 but a discernible trend was that the strength of the spectral signal was inversely correlated with the extent of the deletion suggesting that the intrinsically disordered region contributes to the defined secondary structural elements of the folded C-terminus of PrPC. The calculated secondary structural content of most deletion mutant was comparable to that of WT 23–231 but, uniquely, PrP Δ105–125 showed a reduction in α helix and a concomitant increase in β sheet ().
Figure 3. Monoclonal antibody probing of local structure. (A) The epitope locations of each of 5 MAbs used in the study drawn on a schematic of the prion protein structure, the N-terminus is omitted for clarity. (B) Comparative ELISA of WT 23–231 and Δ105–125 with each of the MAbs shown. Each protein antigen was normalized to 5µM before coating to the plate.
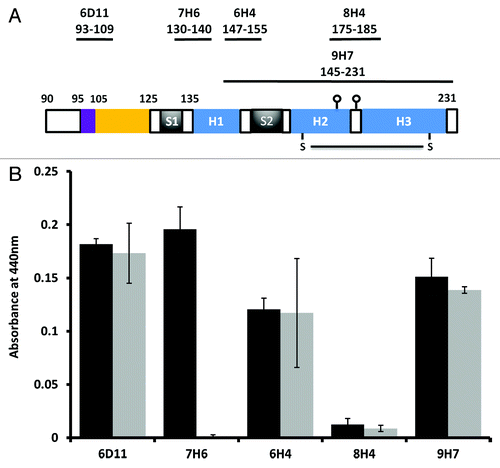
Table 1.CD spectroscopy of purified prion proteins after analysis of α helix and β sheet content using the K2D3 algorithm.
To test whether the unique properties of construct Δ105–125 were dependent on the expression system used, all deletion constructs were also expressed as GFP fusion proteins in insect cells using recombinant baculoviruses.Citation32 Fusion to GFP has been shown to provide a measure of the folding and stability of the tagged protein or protein domain.Citation33,Citation34 Following the construction of recombinant baculoviruses, high titer stocks were used to infect Sf9 cells at high multiplicity and the expression level of each prion-GFP fusion was assessed by western blot with mAb 6H4 (). In addition, GFP fluorescence was measured by flow cytometry and the mean fluorescence plotted after normalization to the intensity of the 6H4 signal. All prion-GFP fusion proteins were expressed at significant levels as detected by western blot and all bar PrP Δ105–125 showed similar levels of fluorescence. However, PrP Δ105–125-GFP showed a lower relative fluorescence than all other constructs including the full-length protein (). When cells expressing WT 23–231-GFP and PrP Δ105–125-GFP were examined at the single cell level, those expressing WT 23–231-GFP fusion showed abundant fluorescence evenly distributed throughout the cytoplasm while those expressing PrP Δ105–125 were more heterogeneous with examples of punctate fluorescence, often sequestered at the poles of the cell (). These data are consistent with PrP Δ105–125 exhibiting an altered degree of stability and routing following expression in insect cells.
Figure 4. Expression of prion deletions in Sf9 cells as fusion proteins with GFP. (A) Western blot of Sf9 cells infected with each prion-GFP fusion with mAb 6H4 at two days post infection. (B) Mean fluorescence of each prion-GFP fusion at 2 d post infection normalized to the 6H4 signal. (C) Fluorescence microscopy of Sf9 cells expressing GFP tagged prion protein. The upper panel is WT 23–231-GFP and the lower is Δ105–125-GFP. The scale bars indicate 400µm.
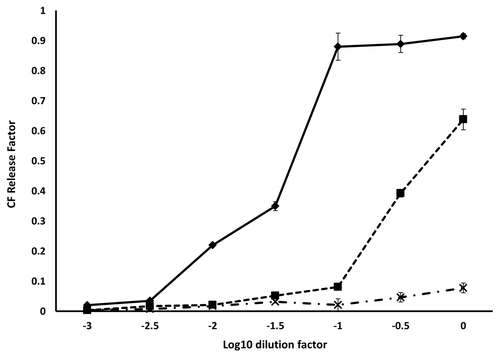
As prion amino acids 105–125 are reported to associate with lipids,Citation17 purified WT 23–231, PrP 90–231 and PrP Δ105–125 proteins were tested for binding to synthetic lipids in vitro. Vesicles formed with 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1'-rac-glycerol) (POPG) or 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (PC) were incubated with each protein and the mixtures subjected to flotation assay. POPG and RNA have been shown previously to be the minimum components necessary for the formation of PrPSc de novo from recombinant PrPC 34 while membranes formed of PC mimic, to some extent, the cytosolic membrane. Gradients were fractionated from the top and the presence of each protein determined by ELISA using mAb 6H4. Both WT 23–231 and PrP 90–231 interacted with POPG as evidenced by their presence in the lower density fractions 1–3 but not with PC vesicles while Δ105–125 showed no binding to either POPG or PC vesicles (). To investigate if membrane binding influenced membrane stability following binding, WT 23–231, PrP 90–231 and PrP Δ105–125 were also incubated with POPG vesicles loaded with carboxyfluorescein. The inclusion of WT 23–231 had a substantial effect on vesicle integrity, causing nearly complete dye release at a concentration of 1µM whereas membrane destabilization by PrP 90–231 required a 10-fold higher concentration () and Δ105–125 showed no membrane destabilization activity, in agreement with the strength of vesicle binding. None of the other internally deleted prion constructs caused dye release consistent with a lack of membrane binding (not shown).
Figure 5. Binding to synthetic lipids. ELISA data for binding of 6H4 to each fraction of a sucrose gradient following flotation assay. Purified prion proteins WT 23–131, 90–231 and Δ105–125 were incubated with POPG (solid lines) or PC (dashed lines) vesicles as described, prior to underloading the gradient. The samples were: WT 23–231 with no lipid (◆), WT 23–231 with PC (■), WT 23–231 with POPG (▲), Δ105–125 with PC (X), Δ105–125 with POPG (□) and 90–231 with POPG (●). 93–231 with PC was not done. Fraction 1 represents the top and fraction 8 the bottom of the gradient. ELISA measurements were performed in triplicate and error bars indicate the standard error mean.
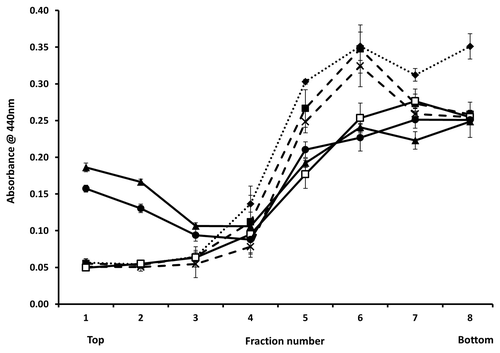
Figure 6. Fluorescence dequenching of CF loaded vesicles by prion protein binding. The initial concentration of each protein was 10 µM. The concentration of lipid remained constant over the dilution series at 250 µg/ml. The samples were: WT 23–231 (◆), 90–231 (■) and Δ105–125 (X). Measurements were performed in triplicate and error bars indicate the standard error mean.
Discussion
Residues in the hydrophobic core of the mouse prion protein are associated with the revelation of direct toxicity in vivoCitation12,Citation13 and in vitro.Citation11 Here, the consequence of deletion of these residues on prion protein structure and function was investigated. Following expression and purification, ANS binding in vitro suggested that deletion of the hydrophobic residues leads to a reduction in solvent accessible binding sites, with the extent of the deletion broadly correlated with the reduction observed, i.e., PrP Δ95–105 displayed the smallest reduction and PrP Δ105–135 the largest. The deleted hydrophobic residues are disordered in the available NMR structuresCitation35,Citation36 and are therefore likely to be solvent accessible. Thus, deletion would be expected to reduce solvent accessibility and ANS signal, broadly as observed. Uniquely however, PrP Δ105–125 was exempt from this trend and demonstrated equivalent fluorescence to WT 23–231, suggesting that Δ105–125 adopts a unique conformation in which ANS binding to alternate residues compensates for those lost by deletion. Interestingly, construct PrP 90–231 also displayed significantly reduced fluorescence suggesting that ANS binding occurs either largely in the region 23–89 or that deletion of amino acids 23–89 stabilizes the folding of residues 90–231, as suggested by the fact that the available structures of PrP solved by high resolution NMRCitation35-Citation37 or X-ray crystallographyCitation38,Citation39 required similar truncations. The ANS fluorescence shown by the internally deleted PrP proteins was reduced in the presence of Cu2+ ions. Cu2+ binds with high affinity in vitro to the octarepeats regionCitation40 and to a further site consisting of His 96 and His 111.Citation21 Copper is also associated with PrPC in vivo.Citation41,Citation42 Reduced ANS binding in the presence of copper would be consistent with reduced solvent accessibility caused by compaction of the tertiary structure, consistent with the in vitro observations that Cu2+ reduces PrPC aggregation and fibrillisation.Citation9 All constructs displayed significantly higher ANS signal at acidic pH, with Δ105–125 displaying the greatest increase compared with WT suggesting that its novel structure may also have an increased sensitivity to pH dependent unfolding. This may be of physiological significance as the protein is normally trafficked to the endosome.Citation27 As with ANS binding at neutrality, increased ANS fluorescence at lower pH was mitigated by the presence of Cu2+ ions.
A novel tertiary structure for Δ105–125 compared with other internally deleted constructs was supported by CD analysis which revealed an altered α helix/β sheet ratio when compared with all other constructs. This is consistent with the data of Thaa et al. who showed that a similar mutant, deleted for residues Δ111–114, had increased β sheet content when compared with WT 23–231.Citation43 Monoclonal antibody binding showed that Δ105–125, unlike WT 23–231, had lost the ability to bind mAb 7H4 despite the retention of the mapped epitope (residues 130–140).Citation29 In the structure of PrP this region encompasses the first β strand, upstream of helix 1, and the observed increase in β sheet content could be due to remodelling of the sheet in this region which obscures the 7H4 epitope. However, confirmation of this would require finer structural probing, for example by twin site ELISAs, to assess local conformation. Most mAb binding to WT 23–231and Δ105–125 was equivalent suggesting little overall change to the protein fold unlike that observed for truncated forms of PrP. 44 Δ105–125 also exhibited singular behavior when tagged with GFP and expressed in in Sf9 cells as a PrP Δ105–125-GFP fusion protein. Fluorescence was reduced overall and had a punctate rather than dispersed pattern, as was shown by WT 23–231. This data are consistent with that of Christensen et al. who concluded that although a PrP mutant lacking residues 95–125 localized to the cytosolic membrane in a variety of mammalian cells, it was prone to increased aggregation when measured by analytical centrifugation.Citation44 The direct toxicity of PrP Δ105–125 in vivo has been suggested to depend on pore formation in cytosolic membranes.Citation16,Citation45 However only PrP 90–231 and WT 23–231 were able to interact with anionic POPG membranes in vitro, consistent with previous studies.Citation17,Citation46 WT 23–231 and PrP 90–231 binding to synthetic membranes also lead to loss of vesicle integrity, although the absence of residues 23–89 reduced the efficiency of membrane destabilization significantly. Prion protein residues 105–125 have been demonstrated to facilitate POPG interactionCitation17 in the context of full length protein, although an isolated peptide consisting of residues 105–126 fails to bind membranes,Citation47 suggesting context dependency in membrane interaction for this sequence. Membrane pore formation as a mechanism of toxicity for PrP Δ105–125 may require the higher complexity of biological membranes.
The unique structural properties of PrP Δ105–125, measured here by a variety of assays, would be consistent with the receptor mediated hypotheses models for prion toxicity. The NMDA receptor has been identified recently as an interaction partner of PrPC,Citation48–Citation51 leading to the speculation that the particular conformation adopted by Δ105–125 is the conformation that binds the NMDA receptor and mediates a toxic signaling outcome. Alternatively, residues 95–125 mediate, in concert with the extreme N terminus residues 23–31 and the length of the intervening sequence, the ability of PrPC to bind to amyloid-β.Citation52 Particular loss of 95–125 could therefore prevent the formation of the specific structure associated with amyloid-β binding and mediate toxicity directly. Further studies of membrane binding and direct interaction with NMDA-R, amyloid-β and PrPSc may help to relate the molecular properties of PrP residues 105–125 with the mechanism of prion associated pathology.
Materials and Methods
All chemical reagents were purchased from Sigma Aldrich unless otherwise stated. Lipids were obtained from Avanti polar lipids. Restriction enzymes were purchased from New England Biolabs (NEB) and used in accordance with manufactures guidelines. Pfu turbo polymerase was purchased from Stratagene and used similarly.
Cloning of deletion mutant constructs
A pET23a vector containing the cloned cDNA of residues 23–231 of murine prion protein was amplified with Pfu Turbo (Stratagene) by outfacing PCR using primers that contained unique terminal HindIII sites. The resultant PCR products were subsequently digested with HindIII, isolated by gel extraction and self-ligated using T7 DNA ligase (NEB). Ligation reactions were transformed into Novablue chemically competent E .coli (Novagen) and plasmid DNA extracted and screened by digestion with HindIII. Putative constructs were verified by sequencing (Source Bioscience). C-terminally GFP tagged constructs were created by explanting each deleted construct in-frame into a pTriEx vector continuing a GFP tag.Citation53
Expression and purification of recombinant prion proteins
Purified recombinant murine prion protein was produced as described.Citation25 Briefly pET23a construct protein expression was induced at 37°C in LB when cultures were at an OD600 of 0.6 using IPTG at 0.2 mM and grown for a further 3hrs. Cells were harvested by centrifugation at 2,500 × g for 20 min at 4°C. Pellets were lysed with lysozyme and solubilised in solubilisation buffer (8 M Urea, 10 mM TRIS-HCl pH 7.5, 1 mM DTT). Expressed protein was purified by affinity chromatography on a 5ml His Trap FF column (GE Healthcare). Eluted peak fractions were pooled and concentrated to 2.5ml by centrifugation at 2,500 × g at 4°C using a Vivaspin concentrator (10,000 MW, Sartorius). Concentrated protein was desalted using PD10 columns (GE Healthcare) pre-equilibrated in solubilisation buffer. By SDS-PAGE gel analysis each recombinant protein was > 99% pure and protein stocks were stored at 4°C.
Refolding of recombinant prion proteins
Purified protein samples denatured in solubilisation buffer were refolded by serial dilution in Chelex-100 (Sigma Aldrich) treated 10mM TRIS-HCl pH 7.5, 1mM DTT. The concentration of each refolded protein was determined using the Beer Lambert equation.
CD spectroscopy
Protein samples were refolded as described and normalized to 0.5mg ml−1. Samples were analyzed under nitrogen using a 1mm path length quartz cuvette in a Chirascan Circular Dichroism Spectrometer (Applied Photophysics UK). Scans were from 200nm to 280nm with 1nm intervals. Four scans per sample were performed at room temperature (21°C) and were normalized to the scan of buffer alone, then averaged.
8-anilino-1-naphthalene sulfonate (ANS) fluorescence
Purified prion protein constructs were refolded as described and adjusted to a concentration of 70µM. 50µl of each protein solution was added to the well of a flat bottomed black 96 well plate (Greiner Germany) to which 50µl of 1.2mM ANS in the same buffer was added and mixed. For Cu2+ containing experiments CuSO4 solution was added to the refolded protein to a cation concentration of 100µM. After incubating for 5 min at room temperature fluorescence was read with a GENios plate reader (Tecan UK) using an excitation wavelength of 360nm and emission of 465nm. Experiments were done in triplicate and results normalized to a control in which ANS was added to appropriate buffer lacking protein.
ELISA
Proteins were refolded as described and normalized to 5µM. Samples were absorbed onto flat bottomed transparent 96 well plates (Nuncalon II) by incubating for 1hr at room temperature in 0.1M NaHCO3. Wells were then washed with TBS and blocked by filling with 5% Casein in TBST and incubating 16hr at 4°C. Wells were then washed with TBS and incubated for 1 h with 50µl of the appropriate primary antibody at 1μg/ml dilution in a solution of 0.5% Casein, TBST. After washing 3 times with TBS, the wells were incubated for 1 h with 50µl of 1:1000 dilution of anti-Mouse IgG peroxidase conjugate (Sigma) in 0.5% casein TBST. Wells were washed 3 times in TBS and 50µl of tetramethylbenzidine (Europa Bioproducts UK) was added to each well. The reaction was stopped by addition of 50µl of 0.1M H2SO4 and the absorbance of each well at 440nm was measured using a GENios plate reader (Tecan UK). Experiments were performed in triplicate. Controls had no protein or no primary antibody added and the absorbance readings were normalized to the reading for no protein.
Western blotting
Protein samples were separated on pre-cast Any kD TRIS-HCl SDS-polyacrylamide gels (Bio-Rad) and transferred onto Immobilon-P transfer membrane (Millipore) using semi dry transfer. Membranes were blocked by incubation in 5% milk powder in TBST (50 mM Tris.HCl pH7.5, 150 mM NaCl, 0.05% Tween 20) for 1 h. at room temperature. The membrane was washed with TBST and incubated with 0.5 μgs/ml mAb 6H4 (Prionics) in a 0.5% casein TBST solution (diluent) for 1 h. After two 10min washes with TBST the blot was incubated with 1:1000 anti-mouse IgG peroxidase conjugate (Sigma) in diluent for 1 h. The membrane was washed twice for 10 min with TBST and the bound conjugate detected using ECL (Amersham Biosciences UK). Band intensities were measured using Quantiscan (BioSoft).
FACS analysis
Recombinant baculoviruses were generated and amplified to high titer stocks as described.Citation53 Spodoptera frugiperda 9 (Sf9) monolayers of 0.9 x 106 cells were infected at an MOI > 3 and incubated for 2 d at 28°C. Cells were removed from the plate and the resultant pellet washed twice before being resuspended in 2.5ml of FACSFlow (Becton Dickenson). Fluorescence was measured using a FACScan (Becton Dickinson).
Fluorescence microscopy
Sf9 cells were infected as described for FACS analysis and GFP fluorescence was observed using an EVOS inverted fluorescence microscope (AMG) at 2 d post infection.
Large Unilamellar Vesicle (LUV) production
Five mg of lipid in chloroform was dried using a mini rotary vacuum for two hrs. The dried lipid films were then re-hydrated in 20mM Tris H.Cl pH 7.4, 100mM NaCl to a concentration of 5 mg ml−1. 1ml of the aqueous lipid suspension was then extruded 10 times using a Lipofast vesicle extruder (Avanti polar lipids) using a 100nm pore poly carbonate membrane resulting in a 100nm vesicle suspension at a concentration of 5 mg ml−1 of lipid. Vesicles were confirmed for size and uniformity by EM. Vesicle stocks were kept at 4°C and used within one week of generation.
Carboxyfluorescein (CF) loaded vesicle production
Vesicles were prepared as before except the rehydration used a buffer of 50mM CF in 20mM Tris.HCl pH7.4. Size exclusion chromatography with Sephadex G-75 in 20mM Tris H.Cl pH 7.4, 100mM NaCl was used to remove excess un-entrapped CF from the CF loaded vesicles. CF loaded vesicles were kept at 4°C and used within one week of generation.
Vesicle binding assay
Refolded prion protein was incubated with POPG or PC lipid vesicles at a protein to lipid ratio of 1:500 at 37°C for 1 h. The protein lipid mixture was then adjusted to 60% sucrose w/v and the protein lipid solution layered under a gradient of 60%:10% sucrose in 20mM Tris H.Cl pH 7.4, 100mM NaCl. Gradients were developed in a TL-100 ultra-centrifuge (Beckman) at 50,000 rpm for three hours at 4°C. The gradients were fractionated from the top and the fractions absorbed to the wells of a Nuncalon II ELISA plate. ELISA was done as described. The experiments were performed in triplicate and signals were normalized to that of a control gradient with protein alone.
CF Vesicle dye release assay
CF loaded vesicles were diluted to 500 µg/ml lipid. 100µl of diluted vesicles was mixed with 100µl of refolded protein in the well of a fluorotrac flat bottomed black plate (Greiner Germany) and incubated for five minutes at room temperature. The fluorescence increase upon incubation with protein was measured using a GENios plate reader (Tecan UK) using excitation and emission wavelengths of 485 nm and 535 nm respectively. Controls of total and no CF dye were measured by incubating 100 µl of diluted vesicles with 100µl of buffer 20mM Tris H.Cl pH7.4, 100 mM NaCl alone (no dye release) or 100 µl of 1% triton X-100 (total dye release). Measurements were performed in triplicate. The fraction of CF dye release at various protein concentrations was calculating by using the equation Rf = (FPro - F0) / (Ftotal - F0) where Rf is the release factor, FPro is the fluorescence reading at a specific protein concentration, F0 is the fluorescence reading for the no dye release control and Ftotal is the fluorescence reading for the total dye release control.
Acknowledgments
A.P. was the grateful recipient of an MRC doctoral training award.
Disclosure of Potential Conflicts of Interest
No potential conflict of interest was disclosed.
References
- Borchelt DR, Rogers M, Stahl N, Telling G, Prusiner SB. Release of the cellular prion protein from cultured cells after loss of its glycoinositol phospholipid anchor. Glycobiology 1993; 3:319 - 29; http://dx.doi.org/10.1093/glycob/3.4.319; PMID: 7691278
- Bocharova OV, Breydo L, Salnikov VV, Baskakov IV. Copper(II) inhibits in vitro conversion of prion protein into amyloid fibrils. Biochemistry 2005; 44:6776 - 87; http://dx.doi.org/10.1021/bi050251q; PMID: 15865423
- Prusiner SB. Prions. Proc Natl Acad Sci U S A 1998; 95:13363 - 83; http://dx.doi.org/10.1073/pnas.95.23.13363; PMID: 9811807
- Hill AF, Collinge J. Subclinical prion infection in humans and animals. Br Med Bull 2003; 66:161 - 70; http://dx.doi.org/10.1093/bmb/66.1.161; PMID: 14522857
- Gambetti P, Dong Z, Yuan J, Xiao X, Zheng M, Alshekhlee A, et al. A novel human disease with abnormal prion protein sensitive to protease. Ann Neurol 2008; 63:697 - 708; http://dx.doi.org/10.1002/ana.21420; PMID: 18571782
- Zou WQ, Puoti G, Xiao X, Yuan J, Qing L, Cali I, et al. Variably protease-sensitive prionopathy: a new sporadic disease of the prion protein. Ann Neurol 2010; 68:162 - 72; http://dx.doi.org/10.1002/ana.22094; PMID: 20695009
- Collinge J, Clarke AR. A general model of prion strains and their pathogenicity. Science 2007; 318:930 - 6; http://dx.doi.org/10.1126/science.1138718; PMID: 17991853
- Simoneau S, Rezaei H, Salès N, Kaiser-Schulz G, Lefebvre-Roque M, Vidal C, et al. In vitro and in vivo neurotoxicity of prion protein oligomers. PLoS Pathog 2007; 3:e125; http://dx.doi.org/10.1371/journal.ppat.0030125; PMID: 17784787
- Baskakov IV, Legname G, Baldwin MA, Prusiner SB, Cohen FE. Pathway complexity of prion protein assembly into amyloid. J Biol Chem 2002; 277:21140 - 8; http://dx.doi.org/10.1074/jbc.M111402200; PMID: 11912192
- Sandberg MK, Al-Doujaily H, Sharps B, Clarke AR, Collinge J. Prion propagation and toxicity in vivo occur in two distinct mechanistic phases. Nature 2011; 470:540 - 2; http://dx.doi.org/10.1038/nature09768; PMID: 21350487
- Forloni G, Angeretti N, Chiesa R, Monzani E, Salmona M, Bugiani O, et al. Neurotoxicity of a prion protein fragment. Nature 1993; 362:543 - 6; http://dx.doi.org/10.1038/362543a0; PMID: 8464494
- Baumann F, Tolnay M, Brabeck C, Pahnke J, Kloz U, Niemann HH, et al. Lethal recessive myelin toxicity of prion protein lacking its central domain. EMBO J 2007; 26:538 - 47; http://dx.doi.org/10.1038/sj.emboj.7601510; PMID: 17245436
- Li A, Christensen HM, Stewart LR, Roth KA, Chiesa R, Harris DA. Neonatal lethality in transgenic mice expressing prion protein with a deletion of residues 105-125. EMBO J 2007; 26:548 - 58; http://dx.doi.org/10.1038/sj.emboj.7601507; PMID: 17245437
- Christensen HM, Harris DA. A deleted prion protein that is neurotoxic in vivo is localized normally in cultured cells. J Neurochem 2009; 108:44 - 56; http://dx.doi.org/10.1111/j.1471-4159.2008.05719.x; PMID: 19046329
- Puoti G, Bizzi A, Forloni G, Safar JG, Tagliavini F, Gambetti P. Sporadic human prion diseases: molecular insights and diagnosis. Lancet Neurol 2012; 11:618 - 28; http://dx.doi.org/10.1016/S1474-4422(12)70063-7; PMID: 22710755
- Solomon IH, Huettner JE, Harris DA. Neurotoxic mutants of the prion protein induce spontaneous ionic currents in cultured cells. J Biol Chem 2010; 285:26719 - 26; http://dx.doi.org/10.1074/jbc.M110.134619; PMID: 20573963
- Wang F, Yin S, Wang X, Zha L, Sy MS, Ma J. Role of the highly conserved middle region of prion protein (PrP) in PrP-lipid interaction. Biochemistry 2010; 49:8169 - 76; http://dx.doi.org/10.1021/bi101146v; PMID: 20718504
- Cardamone M, Puri NK. Spectrofluorimetric assessment of the surface hydrophobicity of proteins. Biochem J 1992; 282:589 - 93; PMID: 1546973
- Swietnicki W, Petersen R, Gambetti P, Surewicz WK. pH-dependent stability and conformation of the recombinant human prion protein PrP(90-231). J Biol Chem 1997; 272:27517 - 20; http://dx.doi.org/10.1074/jbc.272.44.27517; PMID: 9346881
- Uversky VN, Winter S, Löber G. Use of fluorescence decay times of 8-ANS-protein complexes to study the conformational transitions in proteins which unfold through the molten globule state. Biophys Chem 1996; 60:79 - 88; http://dx.doi.org/10.1016/0301-4622(96)00009-9; PMID: 8679928
- Klewpatinond M, Davies P, Bowen S, Brown DR, Viles JH. Deconvoluting the Cu2+ binding modes of full-length prion protein. J Biol Chem 2008; 283:1870 - 81; http://dx.doi.org/10.1074/jbc.M708472200; PMID: 18042548
- Pauly PC, Harris DA. Copper stimulates endocytosis of the prion protein. J Biol Chem 1998; 273:33107 - 10; http://dx.doi.org/10.1074/jbc.273.50.33107; PMID: 9837873
- Taylor DR, Watt NT, Perera WS, Hooper NM. Assigning functions to distinct regions of the N-terminus of the prion protein that are involved in its copper-stimulated, clathrin-dependent endocytosis. J Cell Sci 2005; 118:5141 - 53; http://dx.doi.org/10.1242/jcs.02627; PMID: 16254249
- Thompsett AR, Abdelraheim SR, Daniels M, Brown DR. High affinity binding between copper and full-length prion protein identified by two different techniques. J Biol Chem 2005; 280:42750 - 8; http://dx.doi.org/10.1074/jbc.M506521200; PMID: 16258172
- Wong BS, Vénien-Bryan C, Williamson RA, Burton DR, Gambetti P, Sy MS, et al. Copper refolding of prion protein. Biochem Biophys Res Commun 2000; 276:1217 - 24; http://dx.doi.org/10.1006/bbrc.2000.3604; PMID: 11027613
- Magalhães AC, Silva JA, Lee KS, Martins VR, Prado VF, Ferguson SS, et al. Endocytic intermediates involved with the intracellular trafficking of a fluorescent cellular prion protein. J Biol Chem 2002; 277:33311 - 8; http://dx.doi.org/10.1074/jbc.M203661200; PMID: 12070160
- Rybak SL, Murphy RF. Primary cell cultures from murine kidney and heart differ in endosomal pH. J Cell Physiol 1998; 176:216 - 22; http://dx.doi.org/10.1002/(SICI)1097-4652(199807)176:1<216::AID-JCP23>3.0.CO;2-3; PMID: 9618161
- Pankiewicz J, Prelli F, Sy MS, Kascsak RJ, Kascsak RB, Spinner DS, et al. Clearance and prevention of prion infection in cell culture by anti-PrP antibodies. Eur J Neurosci 2006; 23:2635 - 47; http://dx.doi.org/10.1111/j.1460-9568.2006.04805.x; PMID: 16817866
- Li R, Liu T, Wong BS, Pan T, Morillas M, Swietnicki W, et al. Identification of an epitope in the C terminus of normal prion protein whose expression is modulated by binding events in the N terminus. J Mol Biol 2000; 301:567 - 73; http://dx.doi.org/10.1006/jmbi.2000.3986; PMID: 10966770
- Korth C, Stierli B, Streit P, Moser M, Schaller O, Fischer R, et al. Prion (PrPSc)-specific epitope defined by a monoclonal antibody. Nature 1997; 390:74 - 7; http://dx.doi.org/10.1038/36337; PMID: 9363892
- Louis-Jeune C, Andrade-Navarro MA, Perez-Iratxeta C. Prediction of protein secondary structure from circular dichroism using theoretically derived spectra. Proteins 2011; PMID: 22095872
- Yao Y, Ren J, Jones IM. Amino terminal interaction in the prion protein identified using fusion to green fluorescent protein. J Neurochem 2003; 87:1057 - 65; http://dx.doi.org/10.1046/j.1471-4159.2003.02039.x; PMID: 14622086
- Vasiljevic S, Ren J, Yao Y, Dalton K, Adamson CS, Jones IM. Green fluorescent protein as a reporter of prion protein folding. Virol J 2006; 3:59; http://dx.doi.org/10.1186/1743-422X-3-59; PMID: 16939649
- Wang F, Wang X, Yuan CG, Ma J. Generating a prion with bacterially expressed recombinant prion protein. Science 2010; 327:1132 - 5; http://dx.doi.org/10.1126/science.1183748; PMID: 20110469
- Riek R, Hornemann S, Wider G, Billeter M, Glockshuber R, Wüthrich K. NMR structure of the mouse prion protein domain PrP(121-231). Nature 1996; 382:180 - 2; http://dx.doi.org/10.1038/382180a0; PMID: 8700211
- Riek R, Hornemann S, Wider G, Glockshuber R, Wüthrich K. NMR characterization of the full-length recombinant murine prion protein, mPrP(23-231). FEBS Lett 1997; 413:282 - 8; http://dx.doi.org/10.1016/S0014-5793(97)00920-4; PMID: 9280298
- Zahn R, Liu A, Lührs T, Riek R, von Schroetter C, López García F, et al. NMR solution structure of the human prion protein. Proc Natl Acad Sci U S A 2000; 97:145 - 50; http://dx.doi.org/10.1073/pnas.97.1.145; PMID: 10618385
- Haire LF, Whyte SM, Vasisht N, Gill AC, Verma C, Dodson EJ, et al. The crystal structure of the globular domain of sheep prion protein. J Mol Biol 2004; 336:1175 - 83; http://dx.doi.org/10.1016/j.jmb.2003.12.059; PMID: 15037077
- Knaus KJ, Morillas M, Swietnicki W, Malone M, Surewicz WK, Yee VC. Crystal structure of the human prion protein reveals a mechanism for oligomerization. Nat Struct Biol 2001; 8:770 - 4; http://dx.doi.org/10.1038/nsb0901-770; PMID: 11524679
- Garnett AP, Viles JH. Copper binding to the octarepeats of the prion protein. Affinity, specificity, folding, and cooperativity: insights from circular dichroism. J Biol Chem 2003; 278:6795 - 802; http://dx.doi.org/10.1074/jbc.M209280200; PMID: 12454014
- Kramer ML, Kratzin HD, Schmidt B, Römer A, Windl O, Liemann S, et al. Prion protein binds copper within the physiological concentration range. J Biol Chem 2001; 276:16711 - 9; http://dx.doi.org/10.1074/jbc.M006554200; PMID: 11278306
- Brown DR, Clive C, Haswell SJ. Antioxidant activity related to copper binding of native prion protein. J Neurochem 2001; 76:69 - 76; http://dx.doi.org/10.1046/j.1471-4159.2001.00009.x; PMID: 11145979
- Thaa B, Zahn R, Matthey U, Kroneck PM, Bürkle A, Fritz G. The deletion of amino acids 114-121 in the TM1 domain of mouse prion protein stabilizes its conformation but does not affect the overall structure. Biochim Biophys Acta 2008; 1783:1076 - 84; http://dx.doi.org/10.1016/j.bbamcr.2007.11.007; PMID: 18088603
- Adamson CS, Yao Y, Vasiljevic S, Sy MS, Ren J, Jones IM. Novel single chain antibodies to the prion protein identified by phage display. Virology 2007; 358:166 - 77; http://dx.doi.org/10.1016/j.virol.2006.08.023; PMID: 16996555
- Solomon IH, Biasini E, Harris DA. Ion channels induced by the prion protein: mediators of neurotoxicity. Prion 2012; 6:40 - 5; http://dx.doi.org/10.4161/pri.6.1.18627; PMID: 22453177
- Kazlauskaite J, Sanghera N, Sylvester I, Vénien-Bryan C, Pinheiro TJ. Structural changes of the prion protein in lipid membranes leading to aggregation and fibrillization. Biochemistry 2003; 42:3295 - 304; http://dx.doi.org/10.1021/bi026872q; PMID: 12641461
- Henriques ST, Pattenden LK, Aguilar MI, Castanho MA. PrP(106-126) does not interact with membranes under physiological conditions. Biophys J 2008; 95:1877 - 89; http://dx.doi.org/10.1529/biophysj.108.131458; PMID: 18469080
- Khosravani H, Zhang Y, Tsutsui S, Hameed S, Altier C, Hamid J, et al. Prion protein attenuates excitotoxicity by inhibiting NMDA receptors. J Cell Biol 2008; 181:551 - 65; http://dx.doi.org/10.1083/jcb.200711002; PMID: 18443219
- Stys PK, You H, Zamponi GW. Copper-dependent regulation of NMDA receptors by cellular prion protein: implications for neurodegenerative disorders. J Physiol 2012; 590:1357 - 68; http://dx.doi.org/10.1113/jphysiol.2011.225276; PMID: 22310309
- You H, Tsutsui S, Hameed S, Kannanayakal TJ, Chen L, Xia P, et al. Aβ neurotoxicity depends on interactions between copper ions, prion protein, and N-methyl-D-aspartate receptors. Proc Natl Acad Sci U S A 2012; 109:1737 - 42; http://dx.doi.org/10.1073/pnas.1110789109; PMID: 22307640
- Thellung S, Gatta E, Pellistri F, Corsaro A, Villa V, Vassalli M, et al. Excitotoxicity Through NMDA Receptors Mediates Cerebellar Granule Neuron Apoptosis Induced by Prion Protein 90-231 Fragment. Neurotox Res 2012; 23:301 - 14; http://dx.doi.org/10.1007/s12640-012-9340-9; PMID: 22855343
- Fluharty BR, Biasini E, Stravalaci M, Sclip A, Diomede L, Balducci C, et al. An N-terminal Fragment of the Prion Protein Binds to Amyloid-β Oligomers and Inhibits Their Neurotoxicity in Vivo. J Biol Chem 2013; 288:7857 - 66; http://dx.doi.org/10.1074/jbc.M112.423954; PMID: 23362282
- Pengelley SC, Chapman DC, Mark Abbott W, Lin HH, Huang W, Dalton K, et al. A suite of parallel vectors for baculovirus expression. Protein Expr Purif 2006; 48:173 - 81; http://dx.doi.org/10.1016/j.pep.2006.04.016; PMID: 16797185