Abstract
A significant body of evidence shows that polyglutamine (polyQ) tracts are important for various biological functions. The characteristic polymorphism of polyQ length is thought to play an important role in the adaptation of organisms to their environment. However, proteins with expanded polyQ are prone to form amyloids, which cause diseases in humans and animals and toxicity in yeast. Saccharomyces cerevisiae contain at least 8 proteins which can form heritable amyloids, called prions, and most of them are proteins with glutamine- and asparagine-enriched domains. Yeast prion amyloids are susceptible to fragmentation by the protein disaggregase Hsp104, which allows them to propagate and be transmitted to daughter cells during cell divisions. We have previously shown that interspersion of polyQ domains with some non-glutamine residues stimulates fragmentation of polyQ amyloids in yeast and that yeast prion domains are often enriched in one of these residues. These findings indicate that yeast prion domains may have derived from polyQ tracts via accumulation and amplification of mutations. The same hypothesis may be applied to polyasparagine (polyN) tracts, since they display similar properties to polyQ, such as length polymorphism, amyloid formation and toxicity. We propose that mutations in polyQ/N may be favored by natural selection thus making prion domains likely by-products of the evolution of polyQ/N.
Functional significance of polyQ tracts
PolyQ tracts are common in eukaryotes and are present in 0.5–8% of their proteins depending on the organism, for example, in 0.7, 1.5 and 5% of Homo sapiens, Saccharomyces cerevisiae and Drosophila melonogaster proteins, respectively. This alone seems to hint at an important function and, indeed, polyQ domains were prevalently found in proteins which are involved in transcription regulation, alternative splicing and several other functions and have nuclear localization. It has been demonstrated that polyQ domains are often present in proteins with numerous interaction partners and thus seem to be involved in protein-protein interactions.Citation1
PolyQ tracts can change their length due to DNA repeat instability, which is thought to result from abnormal DNA structures interfering with DNA replication, repair and recombination.Citation2 PolyQ length polymorphism is important for adaption to specific changes in environment and genetic background. For instance, polyQ length variation in the Clock gene has been shown to correlate with breeding date in several bird species.Citation3 Also, variation of polyQ length in the ELF3-encoded protein affects flowering time in Arabidopsis thalianaCitation4 and polyQ length polymorphism in the product of the WC-1 gene affects circadian clock period in Neurospora crassa.Citation5 Similarly, efficiency of the interactions of the human androgen receptor (AR) with coactivators and repressors depends on the length of its polyQ domain which is probably of optimal length in a given genetic background, since changes in its size compromise functionality.Citation6 This indicates that variation in polyQ length may enable quantitative control of various protein activities or interactions and, therefore, can be subject to natural selection.
Role of polyQ in pathology
Proteins with polyQ domains have also been extensively studied due to their role in various diseases, such as Huntington disease, spinocerebellar ataxias, etc. Elongation of the polyQ tract beyond a certain threshold can result in the formation of highly stable, insoluble fibrillar protein aggregates, called amyloids. Notably, longer polyQ tracts are associated with more rapid amyloid formation and faster disease progression.Citation7
Amyloids and possibly other misfolded forms of proteins with expanded polyQ, seem to cause cellular toxicity through a number of mechanisms, including the sequestration of essential cellular proteins. The latter mechanism has been directly demonstrated in a yeast model of Huntington’s disease.Citation8-Citation10 In this model it was shown that toxicity of mutant human huntingtin is caused by the ability of its polymers to induce polymerization of essential cellular glutamine/asparagine (Q/N)-rich proteins and related sequestration of other proteins which interact with these polymers. This mechanism is also likely to be of importance for mammalian polyQ diseases, since amyloids of proteins with polyQ domains can cross-seed the polymerization of numerous mammalian proteins with important functions.Citation11-Citation14 Thus, since elongation of polyQ domains can cause cross-amyloidogenesis of cellular proteins and have a deleterious effect, elongated polyQ should be subject to negative selection.
Yeast prions
S. cerevisiae were found to have numerous proteins which can form amyloids, when produced at high levels.Citation15 However, while most such amyloids disappear upon normalization of the protein level, some of them persist, being inherited in cell divisions. Such amyloids are called yeast prions. Most of the known yeast prion proteins possess domains which are rich in Q and N residues, making them similar to proteins with polyQ and polyN tracts. However, in contrast to amyloids formed by proteins with polyQ and polyN tracts, which can severely slow yeast cell growth at high expression levelsCitation16,Citation17 and cause oxidative stress when produced at more moderate levels,Citation18 prion amyloids do not usually cause overt toxicity.
Yeast prion proteins only rarely convert into amyloid form at their endogenous production levels. For example, the prion form of Sup35, denoted as [PSI+], appears de novo only in ~6 x 10−7 cellsCitation19 in the presence of another prion, [PIN+], which stimulates conversion.Citation20 Low frequency of the de novo appearance of such amyloids seems to be related to the amino acid heterogeneity of yeast prion domains (PrD), since replacement of the first 40 amino acids of the Sup35 PrD with a tract of 62 glutamine residues generated a protein, which was capable of conversion into the prion state with high frequency (~1 x 10−2) at production levels similar to those of Sup35.Citation21 This suggests that amino acid heterogeneity imposes a restriction on the number of ways the PrDs interact in a polymer, thus leading to an in-register interaction, in which identical residues of the PrD in neighboring monomers of a polymer are adjacent to each other.Citation22 There is no such restriction for uniform polyQ sequencesCitation23 and thus, they should be more prone to amyloid formation. Since yeast prion amyloids rarely arise de novo, their continued existence is entirely dependent on their inheritance. This is facilitated by the Hsp104 disaggregase and chaperones from the Hsp70 (Ssa1) and Hsp40 (Sis1) families. These proteins act in concert to fragment prion amyloids, thus increasing the number of prion particles and ensuring prion transmission to daughter cells (reviewed in ref. Citation24). However, despite yeast cells having machinery for prion propagation, the biological significance of yeast prions remains the subject of debate.
PrDs may have originated from PolyQ/N
Recent data obtained in our lab suggest that the susceptibility of prion amyloids to fragmentation is intimately related to the presence of non-Q/N residues in their PrDs. While amyloids formed by proteins with uniform polyQ tracts are inefficiently fragmented in yeast, interspersion of these tracts with tyrosineCitation25 and some other non-glutamine residuesCitation26 stimulates polymer fragmentation. We also noted that yeast PrDs are often rich in only one of these fragmentation-promoting residues (). For example, the Sup35 PrD has ~16% tyrosine, the PrD of Cyc8 has ~20% alanine, while Ure3 and Rnq1 PrDs are rich in serine.
Table 1. Оccurrence of fragmentation-promoting amino acid residues in yeast PrDs
Thus, yeast PrDs can be viewed as polyQ sequences which are highly interspersed with fragmentation-promoting amino acid residues. Such sequences are likely to be less efficient in driving amyloid formation than uniform polyQ tracts, however the resulting amyloids should be prone to fragmentation by Hsp104. These findings allowed us to propose that yeast PrDs may have originated from polyQ by means of mutational substitutions of Q for fragmentation-promoting residues which can then be amplified by repeat expansion. The latter process would enrich the sequence in each case with only one fragmentation-promoting amino acid residue. Since insertions of 9 out of 18 non-Q/N residues into every fifth position of a polyQ domain were found to promote polymer fragmentation,Citation26 the mutated proteins would be rather likely to form fragmentable polymers if they retained the ability to convert into amyloid form. Amplification of areas of polyQ with mutations should result in various types of repeats, which are often present in yeast prions proteins (QA repeats in Cyc8, QG repeats in Rnq1, YNPQGGYQQ repeats in Sup35).
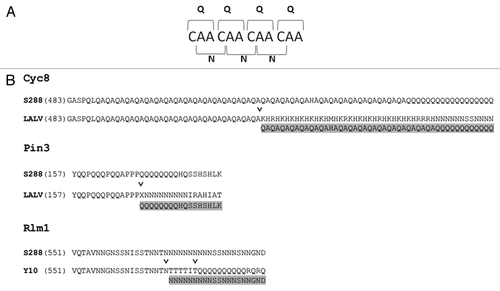
The frequent coincidence of structurally similar Q and N residues in yeast PrDs could be explained by the possibility of interconversion between polyQ and polyN as a result of frameshift mutations in tandem repeats of CAA or ACA triplets, respectively. In support of this possibility, alignments of the amino acid sequences of homologs of several proteins from different strains of S. cerevisiae show that polyN can appear from polyQ (e.g., Cyc8, Pin3) and vice versa (e.g., Rlm1) (). Notably, polyN sequences, similar to polyQ, are common in yeast, display polymorphic length and can drive amyloid formation and toxicity.Citation17 Thus, they may also be able to generate prion-like sequences via the mechanisms described above.
Figure 1. Results of frameshift mutations in polyQ or polyN tracts. (A) frameshift mutations in a polyQ tract encoded by CAA codons result in a tract of polyN. (B) Alignments of portions of the Q/N-rich domains of Cyc8, Pin3 and Rlm1 from various yeast strains. S228, LALV and Y10 denote S228c, LalvinQA23 and Y10 strains of S. cerevisiae. Sequences were taken from the Saccharomyces Genome Database. “˅” depicts the position of deduced frameshift mutations, which convert the sequences of LALV and Y10 proteins into sequences which are nearly identical to the S288 sequence (highlighted in gray).
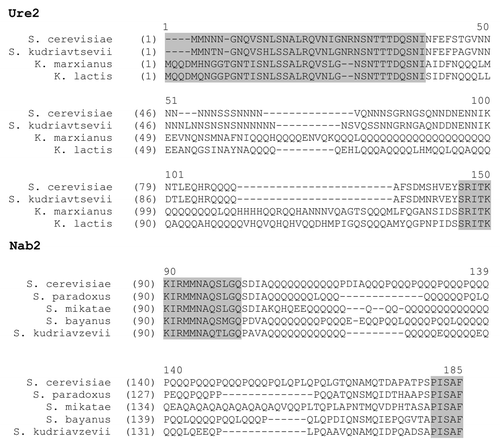
An essential consequence of our hypothesis is that if PrDs derive from polyQ or polyN, they should contain specific traces of being descended from such domains. Such traces can be revealed by comparison of sequences of known PrDs and their homologs from various yeast species (e.g., in Ure2). This analysis demonstrates that such homologs may contain Q/N-rich regions with different patterns of amino acid interspersion in the PrD, indicating that their common ancestor had a uniform polyQ domain which accumulated various interspersions in different lineages (). Many proteins with polyQ and polyN tracts which have not been assayed for their prionogenicity also vary by their interspersion pattern in different yeast species, such as in Nab2 ().
Figure 2. Alignments of homologs of the yeast prionogenic protein Ure2 and Q/N-rich protein Nab2. Conserved sequences are highlighted in gray. Various types of interspersion of polyQ and polyN indicate that the observed sequences are all descended from a polyQ or polyN tract. Similar divergent interspersion patterns can be observed in PrDs and Q/N-rich domains of many other yeast proteins.
An additional important issue for the suggested hypothesis is whether the functional role of a polyQ/N domain is conserved after it has been interspersed with other residues. In general, since a key feature of both polyQ/N and interspersed Q/N-rich sequences is structural disorder, this does not seem implausible. More specifically, the activation domain of the transcription factor Gal4 can be replaced with either a polyQCitation32 or with PrD of Sup35 (our unpublished observation), both of which allow efficient transcription activation, which means that these sequences are more or less equivalent for this function. Also, the prion domains of Sup35 and Ure2, have functions which are unrelated to their prion nature.Citation33-Citation35 Since prion domains are not conserved, these functions are probably not strictly dependent on primary sequence and could have been inherited or evolved from ancestral polyQ/N.
The probability of interspersion of polyQ (and possibly polyN) tracts with non-glutamine residues should be greatly increased if it confers a selective advantage. This is plausible due to the following reasons: (1) mutations within polyQ are known to stabilize polyQ length by lowering repeat length instability which is important because polyQ domains, at least in some cases, seem to have an optimal length for the functioning of their respective proteinsCitation6; (2) appearance and amplification of non-glutamine residues in polyQs is likely to lower the frequency of their conversion into amyloid due to the appearance of a register restriction, which may be advantageous, since most amyloids should be detrimental; (3) interspersion of polyQ with other amino acid residues may decrease toxicity of their amyloids, since such amyloids can be significantly less efficient in amyloid cross-seeding of the essential Q/N-rich protein Sup35, than polyQ amyloids.Citation36
To summarize, interspersion of polyQ or polyN with non-Q/N amino acids is likely to be a common event since it can be favored by natural selection. Interspersion could confer prionogenic properties onto the polyQ/N protein, since interspersing residues can stabilize the monomeric state by decreasing amyloid conversion rates and stimulate fragmentation of amyloid polymers by Hsp104, thus enabling their propagation and inheritance.
Possible scenarios of PrD emergence
All the considerations presented above allow us to speculate how yeast PrDs may have arisen from polyQ or polyN. We propose two scenarios, one that starts with an elongated polyQ/N and another, which starts with a short polyQ/N.
For the first scenario it is important that long polyQ (and possibly N) are prone to further expansion and this should be counterselected due to their increasing propensity for amyloid formation. Mutations in such polyQ/N should be favored due to alleviation of this disadvantage. Once a mutation appears, it can be amplified by the same mechanisms that elongate polyQ/N sequences, turning it into a long heterogenous Q/N-rich sequence.
In the second scenario the process starts from a short, functionally important polyQ/N sequence, which is under selective pressure to expand in response to environmental changes. However the potential advantage of such polyQ/N expansion may be counterbalanced by the disadvantageous aggregation of the expanded protein. In this case, a short polyQ/N sequence may acquire mutations and then expand, thus amplifying these mutations and forming a long heterogenous Q/N-rich domain which is less prone to aggregation and, therefore, not deleterious.
Notably, this hypothesis does not require that the prion properties of proteins be advantageous in their own right. In contrast, we suggest that PrDs could have appeared as a byproduct of mutations in polyQ (or polyN) domains, which means that they are most likely to exist due to their functional activity inherited from polyQ/N. However, if the acquired prion properties are advantageous, as was suggested for Sup35,Citation37 their further evolution could be supported by positive selection.
| Abbreviations: | ||
| polyQ | = | polyglutamine |
| polyN | = | polyasparagine |
| PrD | = | prion domain |
Acknowledgments
The work was funded by grants from the Russian Foundation for Basic Research (12-04-32080 to A.I.A. and 11-04-00442 to M.D.T.-A.), by the Program for Molecular and Cell Biology from the Russian Academy of Science and the Ministry of Education and Science of the Russian Federation. We apologize for not citing all relevant publications due to journal guidelines for the number of references.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Schaefer MH, Wanker EE, Andrade-Navarro MA. Evolution and function of CAG/polyglutamine repeats in protein-protein interaction networks. Nucleic Acids Res 2012; 40:4273 - 87; http://dx.doi.org/10.1093/nar/gks011; PMID: 22287626
- Mirkin SM. Expandable DNA repeats and human disease. Nature 2007; 447:932 - 40; http://dx.doi.org/10.1038/nature05977; PMID: 17581576
- Caprioli M, Ambrosini R, Boncoraglio G, Gatti E, Romano A, Romano M, et al. Clock gene variation is associated with breeding phenology and maybe under directional selection in the migratory barn swallow. PLoS One 2012; 7:e35140; http://dx.doi.org/10.1371/journal.pone.0035140; PMID: 22506071
- Undurraga SF, Press MO, Legendre M, Bujdoso N, Bale J, Wang H, et al. Background-dependent effects of polyglutamine variation in the Arabidopsis thaliana gene ELF3. Proc Natl Acad Sci U S A 2012; 109:19363 - 7; http://dx.doi.org/10.1073/pnas.1211021109; PMID: 23129635
- Michael TP, Park S, Kim TS, Booth J, Byer A, Sun Q, et al. Simple sequence repeats provide a substrate for phenotypic variation in the Neurospora crassa circadian clock. PLoS One 2007; 2:e795; http://dx.doi.org/10.1371/journal.pone.0000795; PMID: 17726525
- Buchanan G, Yang M, Cheong A, Harris JM, Irvine RA, Lambert PF, et al. Structural and functional consequences of glutamine tract variation in the androgen receptor. Hum Mol Genet 2004; 13:1677 - 92; http://dx.doi.org/10.1093/hmg/ddh181; PMID: 15198988
- Chen S, Ferrone FA, Wetzel R. Huntington’s disease age-of-onset linked to polyglutamine aggregation nucleation. Proc Natl Acad Sci U S A 2002; 99:11884 - 9; http://dx.doi.org/10.1073/pnas.182276099; PMID: 12186976
- Kochneva-Pervukhova NV, Alexandrov AI, Ter-Avanesyan MD. Amyloid-mediated sequestration of essential proteins contributes to mutant huntingtin toxicity in yeast. PLoS One 2012; 7:e29832; http://dx.doi.org/10.1371/journal.pone.0029832; PMID: 22253794
- Zhao X, Park YN, Todor H, Moomau C, Masison D, Eisenberg E, et al. Sequestration of Sup35 by aggregates of huntingtin fragments causes toxicity of [PSI+] yeast. J Biol Chem 2012; 287:23346 - 55; http://dx.doi.org/10.1074/jbc.M111.287748; PMID: 22573320
- Gong H, Romanova NV, Allen KD, Chandramowlishwaran P, Gokhale K, Newnam GP, et al. Polyglutamine toxicity is controlled by prion composition and gene dosage in yeast. PLoS Genet 2012; 8:e1002634; http://dx.doi.org/10.1371/journal.pgen.1002634; PMID: 22536159
- Yamanaka T, Tosaki A, Miyazaki H, Kurosawa M, Furukawa Y, Yamada M, et al. Mutant huntingtin fragment selectively suppresses Brn-2 POU domain transcription factor to mediate hypothalamic cell dysfunction. Hum Mol Genet 2010; 19:2099 - 112; http://dx.doi.org/10.1093/hmg/ddq087; PMID: 20185558
- Doi H, Okamura K, Bauer PO, Furukawa Y, Shimizu H, Kurosawa M, et al. RNA-binding protein TLS is a major nuclear aggregate-interacting protein in huntingtin exon 1 with expanded polyglutamine-expressing cells. J Biol Chem 2008; 283:6489 - 500; http://dx.doi.org/10.1074/jbc.M705306200; PMID: 18167354
- Furukawa Y, Kaneko K, Matsumoto G, Kurosawa M, Nukina N. Cross-seeding fibrillation of Q/N-rich proteins offers new pathomechanism of polyglutamine diseases. J Neurosci 2009; 29:5153 - 62; http://dx.doi.org/10.1523/JNEUROSCI.0783-09.2009; PMID: 19386911
- Schaffar G, Breuer P, Boteva R, Behrends C, Tzvetkov N, Strippel N, et al. Cellular toxicity of polyglutamine expansion proteins: mechanism of transcription factor deactivation. Mol Cell 2004; 15:95 - 105; http://dx.doi.org/10.1016/j.molcel.2004.06.029; PMID: 15225551
- Alberti S, Halfmann R, King O, Kapila A, Lindquist S. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell 2009; 137:146 - 58; http://dx.doi.org/10.1016/j.cell.2009.02.044; PMID: 19345193
- Meriin AB, Zhang X, He X, Newnam GP, Chernoff YO, Sherman MY. Huntington toxicity in yeast model depends on polyglutamine aggregation mediated by a prion-like protein Rnq1. J Cell Biol 2002; 157:997 - 1004; http://dx.doi.org/10.1083/jcb.200112104; PMID: 12058016
- Peters TW, Huang M. Protein aggregation and polyasparagine-mediated cellular toxicity in Saccharomyces cerevisiae.. Prion 2007; 1:144 - 53; http://dx.doi.org/10.4161/pri.1.2.4630; PMID: 19164913
- Sorolla MA, Nierga C, Rodríguez-Colman MJ, Reverter-Branchat G, Arenas A, Tamarit J, et al. Sir2 is induced by oxidative stress in a yeast model of Huntington disease and its activation reduces protein aggregation. Arch Biochem Biophys 2011; 510:27 - 34; http://dx.doi.org/10.1016/j.abb.2011.04.002; PMID: 21513696
- Lancaster AK, Bardill JP, True HL, Masel J. The spontaneous appearance rate of the yeast prion [PSI+] and its implications for the evolution of the evolvability properties of the [PSI+] system. Genetics 2010; 184:393 - 400; http://dx.doi.org/10.1534/genetics.109.110213; PMID: 19917766
- Derkatch IL, Bradley ME, Masse SV, Zadorsky SP, Polozkov GV, Inge-Vechtomov SG, et al. Dependence and independence of [PSI(+)] and [PIN(+)]: a two-prion system in yeast?. EMBO J 2000; 19:1942 - 52; http://dx.doi.org/10.1093/emboj/19.9.1942; PMID: 10790361
- Osherovich LZ, Cox BS, Tuite MF, Weissman JS. Dissection and design of yeast prions. PLoS Biol 2004; 2:E86; http://dx.doi.org/10.1371/journal.pbio.0020086; PMID: 15045026
- Ross ED, Edskes HK, Terry MJ, Wickner RB. Primary sequence independence for prion formation. Proc Natl Acad Sci U S A 2005; 102:12825 - 30; http://dx.doi.org/10.1073/pnas.0506136102; PMID: 16123127
- Bugg CW, Isas JM, Fischer T, Patterson PH, Langen R. Structural features and domain organization of huntingtin fibrils. J Biol Chem 2012; 287:31739 - 46; http://dx.doi.org/10.1074/jbc.M112.353839; PMID: 22801429
- Tuite MF, Marchante R, Kushnirov V. Fungal prions: structure, function and propagation. Top Curr Chem 2011; 305:257 - 98; http://dx.doi.org/10.1007/128_2011_172; PMID: 21717344
- Alexandrov IM, Vishnevskaya AB, Ter-Avanesyan MD, Kushnirov VV. Appearance and propagation of polyglutamine-based amyloids in yeast: tyrosine residues enable polymer fragmentation. J Biol Chem 2008; 283:15185 - 92; http://dx.doi.org/10.1074/jbc.M802071200; PMID: 18381282
- Alexandrov AI, Polyanskaya AB, Serpionov GV, Ter-Avanesyan MD, Kushnirov VV. The effects of amino acid composition of glutamine-rich domains on amyloid formation and fragmentation. PLoS One 2012; 7:e46458; http://dx.doi.org/10.1371/journal.pone.0046458; PMID: 23071575
- Wickner RB. [URE3] as an altered URE2 protein: evidence for a prion analog in Saccharomyces cerevisiae.. Science 1994; 264:566 - 9; http://dx.doi.org/10.1126/science.7909170; PMID: 7909170
- Derkatch IL, Bradley ME, Hong JY, Liebman SW. Prions affect the appearance of other prions: the story of [PIN(+)]. [PIN+] Cell 2001; 106:171 - 82; http://dx.doi.org/10.1016/S0092-8674(01)00427-5; PMID: 11511345
- Patel BK, Gavin-Smyth J, Liebman SW. The yeast global transcriptional co-repressor protein Cyc8 can propagate as a prion. Nat Cell Biol 2009; 11:344 - 9; http://dx.doi.org/10.1038/ncb1843; PMID: 19219034
- Rogoza T, Goginashvili A, Rodionova S, Ivanov M, Viktorovskaya O, Rubel A, et al. Non-Mendelian determinant [ISP+] in yeast is a nuclear-residing prion form of the global transcriptional regulator Sfp1. Proc Natl Acad Sci U S A 2010; 107:10573 - 7; http://dx.doi.org/10.1073/pnas.1005949107; PMID: 20498075
- Du Z, Park K-W, Yu H, Fan Q, Li L. Newly identified prion linked to the chromatin-remodeling factor Swi1 in Saccharomyces cerevisiae.. Nat Genet 2008; 40:460 - 5; http://dx.doi.org/10.1038/ng.112; PMID: 18362884
- Gerber HP, Seipel K, Georgiev O, Höfferer M, Hug M, Rusconi S, et al. Transcriptional activation modulated by homopolymeric glutamine and proline stretches. Science 1994; 263:808 - 11; http://dx.doi.org/10.1126/science.8303297; PMID: 8303297
- Hosoda N, Kobayashi T, Uchida N, Funakoshi Y, Kikuchi Y, Hoshino S, et al. Translation termination factor eRF3 mediates mRNA decay through the regulation of deadenylation. J Biol Chem 2003; 278:38287 - 91; http://dx.doi.org/10.1074/jbc.C300300200; PMID: 12923185
- Shewmaker F, Mull L, Nakayashiki T, Masison DC, Wickner RB. Ure2p function is enhanced by its prion domain in Saccharomyces cerevisiae.. Genetics 2007; 176:1557 - 65; http://dx.doi.org/10.1534/genetics.107.074153; PMID: 17507672
- Urakov VN, Valouev IA, Kochneva-Pervukhova NV, Packeiser AN, Vishnevsky AY, Glebov OO, et al. N-terminal region of Saccharomyces cerevisiae eRF3 is essential for the functioning of the eRF1/eRF3 complex beyond translation termination. BMC Mol Biol 2006; 7:34; http://dx.doi.org/10.1186/1471-2199-7-34; PMID: 17034622
- Urakov VN, Vishnevskaya AB, Alexandrov IM, Kushnirov VV, Smirnov VN, Ter-Avanesyan MD. Interdependence of amyloid formation in yeast: implications for polyglutamine disorders and biological functions. Prion 2010; 4:45 - 52; http://dx.doi.org/10.4161/pri.4.1.11074; PMID: 20118659
- Halfmann R, Jarosz DF, Jones SK, Chang A, Lancaster AK, Lindquist S. Prions are a common mechanism for phenotypic inheritance in wild yeasts. Nature 2012; 482:363 - 8; http://dx.doi.org/10.1038/nature10875; PMID: 22337056