Abstract
PrPsc, the pathogenic isoform of PrPc, can convert PrPc into PrPsc through direct interactions. PrPc oligomerization is a required processing step before PrPsc formation, and soluble oligomers appear to be the toxic species in amyloid-related disorders. In the current study, direct interactions between vitamin D2 and human recombinant PrPc (90–231) were observed by Biacore assay, and 3F4 antibody, specific for amino acid fragment 109–112 of PrPc, inhibited this interaction. An ELISA study using3F4 antibody showed that PrPc (101–130), corresponding sequence to human PrP, was affected by vitamin D2, supporting the results of Biacore studies and suggesting that the PrPc sequence around the 3F4 epitope was responsible for the interaction with vitamin D2. Furthermore, the effects of vitamin D2 on disruption of PrPc (90–231) oligomerization were elucidated by dot blot analysis and differential protease k susceptibilities. While many chemical compounds have been proposed as potential therapeutic agents for the treatment of scrapie, most of these are toxic. However, given the safety and blood brain barrier permeability of vitamin D2, we propose that vitamin D2 may be a suitable agent to target PrPc in the brain and therefore is a potential therapeutic candidate for prion disease.
Keywords: :
Introduction
The main event contributing to the pathogenesis of prion disease is the conversion of the cellular prion protein (PrPc) into scrapie prion protein (PrPsc), which is a protease-resistant, insoluble protein. PrPsc is the main component of transmissible amyloid deposits and is essential for progression of the disease.Citation1,Citation2 Prion infectivity can be explained by the direct PrPsc-PrPc interaction.Citation3 In vitro generation of infectious PrPsc has demonstrated the protein-only hypothesis of prion propagation, and the development of a method for the cyclic amplification of PrPsc has provided a highly sensitive assay for the biochemical detection of PrPsc in blood.Citation4,Citation5
Some reports have suggested a role of PrPc in antioxidative defense and have demonstrated the involvement of PrPc in anti-apoptotic pathways.Citation6,Citation7 Moreover, the loss of PrPc leads to amyloid-β production in Alzheimer disease and controls neuroprotective signaling.Citation8 While it has been speculated that the loss of PrPc may contribute to the pathogenesis of prion disease, studies in PrPc-knockout mice have not supported this hypothesis, and the physiological function of PrPc is still unknown.Citation9
Many reports have suggested that the multistep process of conversion from PrPc into PrPsc includes an oligomerization/polymerization step.Citation10,Citation11 The oligomerization or molten-globule state is a preliminary step required for the formation of insoluble protein in the brain, and soluble oligomers appear to be more cytotoxic than mature aggregates.Citation12 The small size of PrPc oligomers facilitates its efficient conversion to the protease k (PK)-resistant form in vitro, which make up most of the components of PrPsc disaggregates that show infectivity.Citation13 Therefore, both PrPsc and PrPc represent potential drug targets for the treatment of related diseases.
Many compounds have shown different efficacies toward the inhibition of aberrant self-assembly of PrPc, dissociation of existing aggregates, protection of cells against neurotoxic effects of the aggregates, and, in some cases, reduction of disease symptoms in vivo; however, there is no curative treatment for prion disease or for the progression of neuronal cell loss in the brain.
One potential therapeutic strategy is to interfere with the direct interaction between PrPc with PrPsc. The β-sheet breaker peptide, which is homologous to the PrP fragments implicated in the abnormal folding, has been shown to partially revert PrPsc to a biochemical and structural state similar to that of PrPc in vitro.Citation14 Recently, cationic tetrapyrrole compound has been shown to display activity toward PrP by binding to a folded domain of human PrP.Citation15 An NMR study demonstrated a direct interaction between PrP and methylene blue on a surface cleft, including a fibrillogenetic region of the protein, and showed that this interaction affected the kinetics of PrP oligomerization, reducing the formation of oligomers.Citation16 Based on a structure-activity relationship study for antiprion activity, researchers demonstrated that tocopherols inhibit prion replication and that this activity can be partially antagonized with rapamycin; these data suggest that signaling pathways of tochopherol targets may interfere with the actions of rapamycin, providing insight into PrP regulation and signaling.Citation17
In the present study, we sought to identify novel compounds that may inhibit prion activity by screening hydrophobic vitamins for their ability to disrupt PrPc oligomerization. Our data demonstrated that vitamin D2 (V-D2) showed a high binding affinity for the truncated form of human recombinant PrPc(90–231) and suppressed PrPc (90–231) oligomerization, resulting in increased susceptibility to PK. This is the first report to suggest the effects of V-D2 on the inhibition of PrPc oligomerization in vitro.
Results
Affinity of V-D2 for Hu-rPrPc (90–231), as measured by Biacore assay
A Biacore assay was employed to determine the affinity of V-D derivatives for Hu-rPrPc (90–231). A strong interaction was observed with V-D2, whereas V-D3 showed no interaction with PrPc (90–231) (). From the sensorgram shown in , we found that the interaction exhibited a high binding affinity, with a KA of 6.17eCitation8 and a KD of 1.62e−9. After saturating PrPc (90–231) with the3F4 antibody, V-D2 binding to PrPc (90–231) was decreased, with a KA of 1.12eCitation8 and a KD of 8.95e−9. () (). The binding of PrPc (101–130) with V-D2 was also decreased after saturating the fragment with the3F4 antibody (), indicating that within the 3F4 epitope, PrPc (90–231) was responsible for the interaction.
Figure 1. Affinity of V-D to PrP, as measured using the Biacore system. (A) The interaction between PrPc (90–231) and V-D2 showed high binding. (B) The interaction between PrPc (90–231) and V-D3 showed no binding affinity. (C) The interaction between PrPc (90–231) and V-D2, after saturating with the3F4 mAb. (D) The interaction between PrPc (101–130) and V-D2, after saturating with the 3F4 mAb. Experiments were performed at least 3 times and similar results were obtained. The figures express the representative results of the experiments.
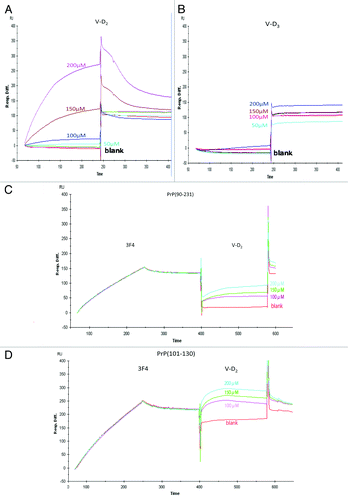
Table 1. Binding kinetics of V-D2 and V-D3 to Hu-rPrP(90–231)
Reactivity of3F4 antibody with Hu-rPrPc (90–231) and PrPc (101–130) bound to V-D2, as monitored by ELISA
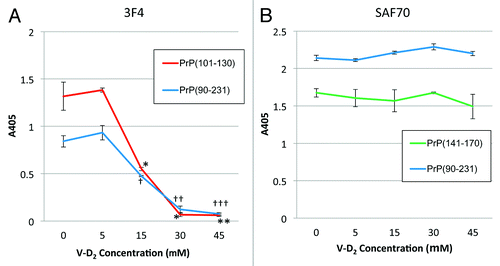
Next, we sought to confirm the responsible fragment within Hu-rPrPc (90–231) that was affected by V-D2. The reactivity of the 3F4 antibody with PrPc (90–231) that was incubated with V-D2 at concentrations ranging from 0 to 45 μM was measured. The 3F4 antibody showed decreasing reactivity toward PrPc (90–231) bound with V-D2 in a dose-dependent manner, and the signal almost reached zero at 30 μM V-D2. The reactivity of 3F4 antibody was similar for PrPc (101–130) incubated with V-D2 (). In the case of this second fragment (101–130), V-D2 reduced the signal by 30% at 15 μM and almost completely abolished the signal at 30 μM. As a control, signals from theSAF70 antibody against both PrPc (90–231) and PrPc (141–170) incubated with V-D2 were not affected by the V-D2 concentration, and the signals were consistently high across all V-D2 concentrations tested ().
Figure 2. Reactivity of mAbs against PrPc with V-D2 by ELISA. (A) The 3F4 epitope on PrPc was affected by V-D2 in a dose-dependent manner. The blue line indicates signals for Hu-rPrPc (90–231), and the red line indicates signals for PrPc (101–130). (B) The SAF70 epitope on PrPc was not affected by V-D2. The blue line indicates signals for Hu-rPrPc (90–231), and the green line indicates signals for PrPc (141–170). Each experiment was performed 3 times by triplicates, and the values express means ± SD *p < 0.006,**p < 0.003 vs. PrPc(101–130) without V-D2, †p < 0.005, ††p < 0.01, †††p < 0.05 vs. Hu-rPrPc(90–231) without V-D2.
Effects of V-D derivatives on PrP oligomerization, as measured by dot blot assay
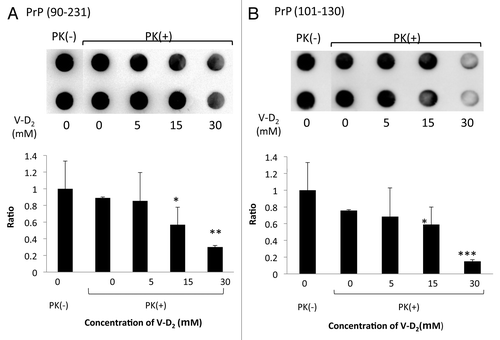
Next, we investigated whether V-D2 could affect the hydrolytic activity of PK using artificial substrates of serine proteases: Boc-Gln-Ala-Arg-MCA for trypsin, Glt-Ala-Ala-Phe-MCA for chymotrypsin, and Suc-Ala-Ala-Ala-MCA for elastase. As expected, V-D2 did not affect the activity of PK toward these substrates (data not shown). Thus, we studied the effects of V-D2 on the formation of Hu-rPrPc (90–231) and PrPc (101–130) oligomers by measuring the protein remaining on the membrane before and after PK digestion of PrPc, detected as a signal from 3F4 antibody. Signals from the spots of PrPc (90–231) were decreased with increasing concentrations of V-D2, indicating the V-D2 increased the susceptibility of PrPc (90–231) to PK (). Similarly, increasing concentrations of V-D2 also caused decreased signals for PrPc (101–130) (). Calculation of the average pixel density of each spot demonstrated that the percentage decrease in the average pixel density was 70% for PrPc (90–231) and 85% for PrPc (101–130) when incubated with 30 μM V-D2.
Figure 3. Sensitivity of PrPc to protease K following incubation with V-D2. V-D2 increased the sensitivity of Hu-rPrPc (90–231) (A) and PrPc (101–130) (B) to PK in a dose-dependent manner. The average pixel density of each spot was measured by NIH image analysis after subtracting the mean background pixel density from that of the test spots. Experiments were performed 3 times by duplicates, and the representative results were expressed. Values are means ± SD of three experiments. *p < 0.05, **p < 0.02, ***p < 0.01 vs. Hu-rPrPc (90–231) or PrPc (101–130) with PK in the absence of V-D2.
Discussion
PrPc is a sialoglycoprotein with a molecular weight of approximately 33–35 kDa that is expressed predominantly in neurons. Studies have shown that fragments of PrPc exhibit a variety of different phenotypes. For example, PrPc (90–231), the N-terminal truncated fragment of PrPc, corresponds to the core of the PK-resistant prion protein, with similar pathogenic features as PrPsc.Citation18,Citation19
Gerstmann-Straussler-Sceinker disease and Creutzfeldt-Jacob disease are well-known naturally occurring prion disease in human, which are caused by mutations in the PrP geneCitation20,Citation21 and the mutations directly link to conformational conversion from PrPc to PrPsc and amplification of PrPsc without exogenous PrPsc.Citation21,Citation22 The N-terminal of PrP(90–231) is highly flexible, and the region of PrP(90–112) lacks defined secondary structure and is thought as a loop or hinge region, which contains naturally occurring mutations region, and could be destabilized by mutations and induce the conformational transition and multimerization of PrPc.Citation23-Citation26 A recent report provides the evidence that mutations between residues 101–112 play a critical role in enhancing multimerization and spontaneous aggregation of prion protein using phage-display system.Citation27
The PrP (89–112) fragment has been also reported to selectively bind to native PrPsc during prion infection, and the 3F4 antibody could recognize the sequence.Citation28 The PrPc (106–126) fragment is highly conserved among various species and is thought to be one of the key domains of PrP involved in amyloid formation.Citation29,Citation30 Moreover, PrPc (106–126) has similar physicochemical and pathological properties as PrPsc, exhibits a high intrinsic ability to polymerize into amyloid-like fibrils, shows resistance to PK, and is neurotoxic in vitro.Citation31 Another fragment, PrP (118–135), a putative transmembrane domain, induces apoptotic neuronal cell death in rat cortical neurons independently of its aggregation.Citation32
Thus, the N-terminal of PrP(90–231) is the therapeutic target in prion diseae. In the present study, we used the PrPc (101–130) fragment, which is completely preserved between human and hamster prion protein, and it contains the PrP(101–112) and a transmembrane domain.
We conducted Biacore assays to examine the binding of PrPc fragments with V-D2. Our data suggested a strong interaction between Hu-rPrPc (90–231) and V-D2, and presaturation of Hu-rPrPc (90–231) or PrPc (101–130) with the 3F4 antibody inhibited the binding of V-D2 to PrPc, indicating that the PrPc sequence containing the 3F4 epitope was involved in binding to V-D2. The 3F4 epitope in PrP is a flexible region as a component of the conformational rearrangement and it participates in the conformational changes from PrPc to PrPsc,Citation33 and V-D2 could inhibit the changes. Additionally, our ELISA study supported the Biacore data, and PrPc (101–130), which possesses the 3F4 epitope, contained the sequence required for the interaction with V-D2.
Several techniques to detect the presence of PrPc (90–231) oligomers have been proposed, and the most common technique is SDS-PAGE analysis followed by western blotting using anti-PrP mAbs.Citation34 The PrPc (90–231) oligomer is sensitive to SDS, and exposure to SDS causes dissociation of the oligomer, resulting in no detection of changes in oligomerization by western blot analysis (data not shown). Therefore, in the present study, we employed dot blotanalysis because this method has allows for the evaluation of oligomer formation without SDS contamination in the samples. Our data indicated that the PK susceptibility of Hu-rPrPc (90–231) was increased by V-D2, suggesting that Hu-rPrPc (90–231) oligomers dissociated upon binding to V-D2. Furthermore, we are studying the potential effects of V-D2 to inhibit the conformational transition from α-helix to β-sheet of PrPc.
V-D is classified as a secosteroid and has 2 distinctive forms: V-D2 and V-D3. V-D3 is a 27-carbon molecule derivative of cholesterol, and V-D2 is a 28-carbon molecule derived from plant sterol ergosterol that contains a double bond between carbons 22 and 23. Due to its cis-triene structure, V-D is susceptible to oxidation, UV light-induced conformational changes, heat-induced conformational changes, and attack by free radicals.Citation35 V-D2 and V-D3 appear to have similar biological effects in humans, and V-D3 is about 4 times as potent as V-D2.Citation36-Citation38 However, we observed differences in the interactions of V-D2 and V-D3 with PrPc (90–231). Interestingly, V-D2 showed higher affinity for Hu-rPrPc (90–231), while V-D3 showed no affinity for this fragment.
Interestingly, V-D2 is manufactured by exposing a fat extract of yeast to UV light, and no metabolites of V-D2 are normally detectable in the blood of humans or primates, unless administered from an external source.Citation38,Citation39 Thus, V-D2 is not a physiological product and is instead regarded as a drug; the metabolites generated for V-D2 are not equivalent to those for V-D3.Citation40 In contrast to V-D2, V-D3 is the natural metabolite generated within the skin and oils of fur, and V-D3 is a substrate for both microsomal and mitochondrial 25-hydroxylases, which do not act on V-D2.Citation39,Citation41,Citation42 Furthermore, the V-D binding protein shows lower affinity for V-D2 than V-D3 and its metabolites.Citation43
The only structural difference between V-D2 and V-D3 is the presence or absence of the C24 methyl group and C22–C23 double bond in the side chains. In general, the presence of a double bond in a linear structure is known to have an influence on the conformational flexibility of the molecule through allylic strain and the rigidity of the double bond against rotation.Citation44,Citation45 Therefore, we hypothesize that the conformational restriction arising from the presence of the double bond in the side chain of V-D2 facilitated the binding of V-D2 with the recognition site of PrPc (90–231).
Contemporary views categorize V-D as a prosteroid hormone rather than a vitamin, and V-D has been shown to have immunomodulatory properties, affecting a reduction in pro-inflammatory immune pathways.Citation46,Citation47 Moreover, V-D has been shown to permeate the blood-brain barrier and to exert beneficial effects in disease progression, reducing the relapse risk in multiple sclerosis through its immunoregulatory effects.Citation48 Therefore, our data demonstrating the role of V-D2 in the regulation of prion oligomerization have implications for the treatments of a variety of prion-related diseases.
In conclusion, our data suggested that V-D2 can bind directly with the region flanking the residues109–112 in prion protein, which could act as a loop or hinge region critical to PrP’s conformational transitions and multimerization. Therefore, V-D2, which is safe for consumption and administration in humans, may be useful for inhibiting the direct interaction between PrPc and PrPsc in clinical setting.
Materials and Methods
PrPc samples of Human recombinant PrPc (90–231)(Hu-rPrP[90–231]) were obtained from Jena Bioscience (PR-911), and synthetic peptides of PrPc (101–130), and PrPc (141–170), both peptides correspond to human PrP sequences, were obtained from OPERON Biotechnologies. Monoclonal antibody (mAb) 3F4 specific for PrPc (109–112) fragment and mAb SAF70 specific for PrPc (142–160) fragment were obtained from Sigma (P1115) and SPIbio (A03206) respectively. V-D derivatives, including V-D2 and V-D3, were from Cayman Chemical (11791, 11792, 9000683, 71820), and other chemical compounds, including PK and phenylmethylsulfonyl fluoride (PMSF) were obtained from Nacalai Tesque (04130-06, 27327-81). The substrate for ELISA detection, p-nitrophenyl phosphate, was from Sigma (N9389-50TAB). All PrP samples and V-D derivatives were dissolved in minimal amount of DMSO and diluted in Tris-buffered saline (TBS; 20 mM Tris, 34 mM NaCl, pH 7.4) at the indicated concentrations.
Biacore analysis
We used a surface plasmon resonance (SPR)-based Biacore 3000 system (GE Healthcare) to analyze the molecular interactions between PrPc and V-D derivatives.Citation49 The Hu-rPrPc (90–231) fragment (200 μg/mL) was covalently linked to a sensor chip CM5 (carboxymethylated dextran surface) (BR-1003-99) through the use of amine-coupling chemistry.Citation50 Solutions of V-D derivatives were injected over the surface at 25°C with a flow rate of 20 μL/min in HBS running buffer (10 mM Hepes, pH 7.4, 150 mM NaCl, 3 mM EDTA, 0.005% [v/v] surfactant P20). After injection, analyte solutions were replaced by HBS at a continuous flow rate of 20 μL/min. Surface regeneration was accomplished by injecting 10 mM glycine-HCl (pH 1.5; 1 min contact time). All analyte solutions were run simultaneously over a control flow cell containing a blank surface (with no immobilized protein). Each sensorgram (time course of the SPR signal) was subtracted for the response observed in the control flow cell and normalized to a baseline of 0 resonance units. To determine the stoichiometry of the interaction between Hu-rPrPc (90–231) and V-D derivatives, various concentrations (20, 50, 100, 150, and 200 μM) of V-D derivatives were passed for 3 min over the sensor chip with immobilized Hu-rPrPc (90–231) with 60-μL injections at a flow rate of 20 μL/min to obtain a saturating response. Kinetic data were interpreted with a simple 1:1 binding model, and interaction rate constants were calculated using BIAevaluation 4.1 SPR Kinetics software (GE Healthcare). We also studied the interactions of Hu-rPrPc (90–231) and PrPc (101–130) with V-D2 after achieving a saturating response for 3F4 at 2 μg/mL with 60-μL injections at a flow rate of 20 μL/min.
ELISA
Microplate wells were coated with 50 μL of Hu- rPrPc (90–231), PrPc (101–130), or PrPc (141–170), at the concentration of 8 μg/mL, overnight at 4°C. After removal of excess sample, the sample in each well was incubated with 50 μL V-D2 or V-D3 at the indicated concentrations (0, 5, 15, 30, or 45 μM) for another 12 h at 37°C. After removal of the V-D derivative solution, the wells were incubated with TBS containing 3% bovine serum albumin (BSA), pH 7.4, overnight at 4°C, washed with TBST (TBS containing 0.05% Tween-20, pH 7.4), and incubated for an additional 2 h at 37°C with 50 μL of one of several mAbs against PrPc (1:1000 dilution) in TBS containing 1% BSA (pH 7.4) as the primary antibody. After incubation, the wells were washed with TBST and incubated for 1 h at 37°C with 50 μL anti-mouse IgG-AP (Promega, S372B) as the secondary antibody. After incubation, the wells were washed with TBST, and bound antibody was detected by addition of p-nitrophenyl phosphate and measured at 405 nm using a spectrophotometric plate reader (Molecular Device, model 680) after several hours, depending on the titer of each mAbs. All washing steps were performed 3 times.
Dot blot assay
Hu-rPrPc (90–231) or PrPc (101–130) (8 μg/mL) were mixed with V-D2 at the indicated concentrations (0, 5, 15, or 30 μM) for 12 h at 37°C in Eppendorf tubes. Then, 100 μL of each suspension was applied to a DP48 dot plate (Advantec, FLE 348AA) and blotted onto a methanol-immersed polyvinylidene difluoride (PVDF) membrane (0.2 μm; Life Technologies, LC2002) with absorption by vacuum pump. The PVDF membrane was then removed, thoroughly air-dried, and rinsed in TBS. The PVDF membrane was incubated with 5 mL PK solution (8 μg/mL in TBS, pH 7.4) for 90 min at 37°C with constant shaking. After removal of the PK solution, the reaction was stopped by washing the membrane with TBS followed by incubation with 3 mM PMSF at room temperature for 30 min and rinsing twice with TBS. The membrane was subsequently processed as a standard western blot using 3F4 mAb as a primary antibody and anti-mouse IgG-HRP (GE Healthcare, NA931V) as a secondary antibody to detect the protein remaining after PK digestion. Protein remaining on the PVDF membrane was treated with Immobilon Western Chemiluminescent HRP Substrate (Millipore, WBKLS0100) and detected by LAS-3000 (Fuji Film).
| Abbreviations: | ||
| PrPc | = | cellular prion protein |
| PrPsc | = | scrapie prion protein |
| PK | = | protease k |
| V-D | = | vitamin D |
| SPR | = | surface plasmon resonance |
Acknowledgments
We would like to thank Dr. Hiroshi Imagawa and Dr. Hironobu Takahasi for their helpful discussions and comments about the manuscript. We would also like to thank Editage for providing editorial assistance.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Prusiner SB. Novel proteinaceous infectious particles cause scrapie. Science 1982; 216:136 - 44; http://dx.doi.org/10.1126/science.6801762; PMID: 6801762
- Prusiner SB. Prions. Proc Natl Acad Sci U S A 1998; 95:13363 - 83; http://dx.doi.org/10.1073/pnas.95.23.13363; PMID: 9811807
- Cohen FE, Pan KM, Huang Z, Baldwin M, Fletterick RJ, Prusiner SB. Structural clues to prion replication. Science 1994; 264:530 - 1; http://dx.doi.org/10.1126/science.7909169; PMID: 7909169
- Castilla J, Saá P, Hetz C, Soto C. In vitro generation of infectious scrapie prions. Cell 2005; 121:195 - 206; http://dx.doi.org/10.1016/j.cell.2005.02.011; PMID: 15851027
- Castilla J, Saá P, Soto C. Detection of prions in blood. Nat Med 2005; 11:982 - 5; PMID: 16127436
- Brown DR, Wong BS, Hafiz F, Clive C, Haswell SJ, Jones IM. Normal prion protein has an activity like that of superoxide dismutase. Biochem J 1999; 344:1 - 5; http://dx.doi.org/10.1042/0264-6021:3440001; PMID: 10548526
- Kuwahara C, Takeuchi AM, Nishimura T, Haraguchi K, Kubosaki A, Matsumoto Y, et al. Prions prevent neuronal cell-line death. Nature 1999; 400:225 - 6; http://dx.doi.org/10.1038/22241; PMID: 10421360
- Nieznanski K, Choi JK, Chen S, Surewicz K, Surewicz WK. Soluble prion protein inhibits amyloid-β (Aβ) fibrillization and toxicity. J Biol Chem 2012; 287:33104 - 8; http://dx.doi.org/10.1074/jbc.C112.400614; PMID: 22915585
- Hetz C, Maundrell K, Soto C. Is loss of function of the prion protein the cause of prion disorders?. Trends Mol Med 2003; 9:237 - 43; http://dx.doi.org/10.1016/S1471-4914(03)00069-8; PMID: 12829011
- Rezaei H. Prion protein oligomerization. Curr Alzheimer Res 2008; 5:572 - 8; http://dx.doi.org/10.2174/156720508786898497; PMID: 19075584
- Kelly JW. The alternative conformations of amyloidogenic proteins and their multi-step assembly pathways. Curr Opin Struct Biol 1998; 8:101 - 6; http://dx.doi.org/10.1016/S0959-440X(98)80016-X; PMID: 9519302
- Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem 2006; 75:333 - 66; http://dx.doi.org/10.1146/annurev.biochem.75.101304.123901; PMID: 16756495
- Silveira JR, Raymond GJ, Hughson AG, Race RE, Sim VL, Hayes SF, et al. The most infectious prion protein particles. Nature 2005; 437:257 - 61; http://dx.doi.org/10.1038/nature03989; PMID: 16148934
- Soto C, Kascsak RJ, Saborío GP, Aucouturier P, Wisniewski T, Prelli F, et al. Reversion of prion protein conformational changes by synthetic beta-sheet breaker peptides. Lancet 2000; 355:192 - 7; http://dx.doi.org/10.1016/S0140-6736(99)11419-3; PMID: 10675119
- Vidal C, Herzog C, Haeberle AM, Bombarde C, Miquel MC, Carimalo J, et al. Early dysfunction of central 5-HT system in a murine model of bovine spongiform encephalopathy. Neuroscience 2009; 160:731 - 43; http://dx.doi.org/10.1016/j.neuroscience.2009.02.072; PMID: 19285121
- Cavaliere P, Torrent J, Prigent S, Granata V, Pauwels K, Pastore A, et al. Binding of methylene blue to a surface cleft inhibits the oligomerization and fibrillization of prion protein. Biochim Biophys Acta 2013; 1832:20 - 8; http://dx.doi.org/10.1016/j.bbadis.2012.09.005; PMID: 23022479
- Muyrers J, Klingenstein R, Stitz L, Korth C. Structure-activity relationship of tocopherol derivatives suggesting a novel non-antioxidant mechanism in antiprion potency. Neurosci Lett 2010; 469:122 - 6; http://dx.doi.org/10.1016/j.neulet.2009.11.057; PMID: 19945507
- James TL, Liu H, Ulyanov NB, Farr-Jones S, Zhang H, Donne DG, et al. Solution structure of a 142-residue recombinant prion protein corresponding to the infectious fragment of the scrapie isoform. Proc Natl Acad Sci U S A 1997; 94:10086 - 91; http://dx.doi.org/10.1073/pnas.94.19.10086; PMID: 9294167
- Liu H, Farr-Jones S, Ulyanov NB, Llinas M, Marqusee S, Groth D, et al. Solution structure of Syrian hamster prion protein rPrP(90-231). Biochemistry 1999; 38:5362 - 77; http://dx.doi.org/10.1021/bi982878x; PMID: 10220323
- Prusiner SB. Molecular biology and pathogenesis of prion diseases. Trends Biochem Sci 1996; 21:482 - 7; http://dx.doi.org/10.1016/S0968-0004(96)10063-3; PMID: 9009832
- Collinge J. Human prion diseases and bovine spongiform encephalopathy (BSE). Hum Mol Genet 1997; 6:1699 - 705; http://dx.doi.org/10.1093/hmg/6.10.1699; PMID: 9300662
- Prusiner SB, Scott MR, DeArmond SJ, Cohen FE. Prion protein biology. Cell 1998; 93:337 - 48; http://dx.doi.org/10.1016/S0092-8674(00)81163-0; PMID: 9590169
- James TL, Liu H, Ulyanov NB, Farr-Jones S, Zhang H, Donne DG, et al. Solution structure of a 142-residue recombinant prion protein corresponding to the infectious fragment of the scrapie isoform. Proc Natl Acad Sci U S A 1997; 94:10086 - 91; http://dx.doi.org/10.1073/pnas.94.19.10086; PMID: 9294167
- Liu H, Farr-Jones S, Ulyanov NB, Llinas M, Marqusee S, Groth D, et al. Solution structure of Syrian hamster prion protein rPrP(90-231). Biochemistry 1999; 38:5362 - 77; http://dx.doi.org/10.1021/bi982878x; PMID: 10220323
- Huang Z, Gabriel JM, Baldwin MA, Fletterick RJ, Prusiner SB, Cohen FE. Proposed three-dimensional structure for the cellular prion protein. Proc Natl Acad Sci U S A 1994; 91:7139 - 43; http://dx.doi.org/10.1073/pnas.91.15.7139; PMID: 7913747
- Leclerc E, Peretz D, Ball H, Sakurai H, Legname G, Serban A, et al. Immobilized prion protein undergoes spontaneous rearrangement to a conformation having features in common with the infectious form. EMBO J 2001; 20:1547 - 54; http://dx.doi.org/10.1093/emboj/20.7.1547; PMID: 11285219
- Verma A, Sharma S, Ganguly NK, Majumdar S, Guptasarma P, Luthra-Guptasarma M. Identification and characterization of a spontaneously aggregating amyloid-forming variant of human PrP((90-231)) through phage-display screening of variants randomized between residues 101 and 112. Int J Biochem Cell Biol 2008; 40:663 - 76; http://dx.doi.org/10.1016/j.biocel.2007.10.009; PMID: 18023239
- Moroncini G, Kanu N, Solforosi L, Abalos G, Telling GC, Head M, et al. Motif-grafted antibodies containing the replicative interface of cellular PrP are specific for PrPSc. Proc Natl Acad Sci U S A 2004; 101:10404 - 9; http://dx.doi.org/10.1073/pnas.0403522101; PMID: 15240877
- Hanan E, Priola SA, Solomon B. Antiaggregating antibody raised against human PrP 106-126 recognizes pathological and normal isoforms of the whole prion protein. Cell Mol Neurobiol 2001; 21:693 - 703; http://dx.doi.org/10.1023/A:1015199904354; PMID: 12043842
- Florio T, Paludi D, Villa V, Principe DR, Corsaro A, Millo E, et al. Contribution of two conserved glycine residues to fibrillogenesis of the 106-126 prion protein fragment. Evidence that a soluble variant of the 106-126 peptide is neurotoxic. J Neurochem 2003; 85:62 - 72; http://dx.doi.org/10.1046/j.1471-4159.2003.01664.x; PMID: 12641727
- Brown DR, Schmidt B, Kretzschmar HA. Role of microglia and host prion protein in neurotoxicity of a prion protein fragment. Nature 1996; 380:345 - 7; http://dx.doi.org/10.1038/380345a0; PMID: 8598929
- Chabry J, Ratsimanohatra C, Sponne I, Elena PP, Vincent JP, Pillot T. In vivo and in vitro neurotoxicity of the human prion protein (PrP) fragment P118-135 independently of PrP expression. J Neurosci 2003; 23:462 - 9; PMID: 12533606
- Kanyo ZF, Pan KM, Williamson RA, Burton DR, Prusiner SB, Fletterick RJ, et al. Antibody binding defines a structure for an epitope that participates in the PrPC-->PrPSc conformational change. J Mol Biol 1999; 293:855 - 63; http://dx.doi.org/10.1006/jmbi.1999.3193; PMID: 10543972
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970; 227:680 - 5; http://dx.doi.org/10.1038/227680a0; PMID: 5432063
- Blackwood JE, Gladys CL, Loening KI, Petrarca AE, Ruth JE. Unambiguous specification of stereoisomerism about a double bonds. J Am Chem Soc 1968; 90:509 - 10; http://dx.doi.org/10.1021/ja01004a063
- Hunt RD, Garcia FG, Hegsted DM, Kaplinsky N. Vitamin D2 and vitamin D3 in new world primates: influence on calcium absorption. Science 1967; 158:943 - 5; http://dx.doi.org/10.1126/science.157.3791.943
- Steenbock H, Kletzien SWF, Haplin JG. The reaction of the chicken to irradiated ergosterol and irradiated yeast as contrasted with natural vitamin D of fish liver oils. J Biol Chem 1932; 97:249 - 64
- Trang HM, Cole DE, Rubin LA, Pierratos A, Siu S, Vieth R. Evidence that vitamin D3 increases serum 25-hydroxyvitamin D more efficiently than does vitamin D2. Am J Clin Nutr 1998; 68:854 - 8; PMID: 9771862
- Marx SJ, Jones G, Weinstein RS, Chrousos GP, Renquist DM. Differences in mineral metabolism among nonhuman primates receiving diets with only vitamin D3 or only vitamin D2. J Clin Endocrinol Metab 1989; 69:1282 - 90; http://dx.doi.org/10.1210/jcem-69-6-1282; PMID: 2555385
- Mawer EB, Jones G, Davies M, Still PE, Byford V, Schroeder NJ, et al. Unique 24-hydroxylated metabolites represent a significant pathway of metabolism of vitamin D2 in humans: 24-hydroxyvitamin D2 and 1,24-dihydroxyvitamin D2 detectable in human serum. J Clin Endocrinol Metab 1998; 83:2156 - 66; http://dx.doi.org/10.1210/jc.83.6.2156; PMID: 9626155
- Holmberg I, Berlin T, Ewerth S, Björkhem I. 25-Hydroxylase activity in subcellular fractions from human liver. Evidence for different rates of mitochondrial hydroxylation of vitamin D2 and D3. Scand J Clin Lab Invest 1986; 46:785 - 90; http://dx.doi.org/10.3109/00365518609084051; PMID: 3026027
- Guo YD, Strugnell S, Back DW, Jones G. Transfected human liver cytochrome P-450 hydroxylates vitamin D analogs at different side-chain positions. Proc Natl Acad Sci U S A 1993; 90:8668 - 72; http://dx.doi.org/10.1073/pnas.90.18.8668; PMID: 7690968
- Jones G, Byrnes B, Palma F, Segev D, Mazur Y. Displacement potency of vitamin D2 analogs in competitive protein-binding assays for 25-hydroxyvitamin D3, 24,25-dihydroxyvitamin D3, and 1,25-dihydroxyvitamin D3. J Clin Endocrinol Metab 1980; 50:773 - 5; http://dx.doi.org/10.1210/jcem-50-4-773; PMID: 6965943
- Hoffmann RW. Allylic 1,3-strain as a controlling factor in stereoselective Transformations. Chem Rev 1989; 89:1841 - 60; http://dx.doi.org/10.1021/cr00098a009
- Wiberg KB, Martin E. Barriers to rotation adjacent to double bonds. J Am Chem Soc 1985; 107:5035 - 41; http://dx.doi.org/10.1021/ja00304a002; PMID: 22175333
- Tsoukas CD, Provvedini DM, Manolagas SC. 1,25-dihydroxyvitamin D3: a novel immunoregulatory hormone. Science 1984; 224:1438 - 40; http://dx.doi.org/10.1126/science.6427926; PMID: 6427926
- Lemire JM. Immunomodulatory actions of 1,25-dihydroxyvitamin D3. J Steroid Biochem Mol Biol 1995; 53:599 - 602; http://dx.doi.org/10.1016/0960-0760(95)00106-A; PMID: 7626516
- Simpson S Jr., Taylor B, Blizzard L, Ponsonby AL, Pittas F, Tremlett H, et al. Higher 25-hydroxyvitamin D is associated with lower relapse risk in multiple sclerosis. Ann Neurol 2010; 68:193 - 203; PMID: 20695012
- Malmqvist M. BIACORE: an affinity biosensor system for characterization of biomolecular interactions. Biochem Soc Trans 1999; 27:335 - 40; PMID: 10093759
- Johnsson B, Löfås S, Lindquist G. Immobilization of proteins to a carboxymethyldextran-modified gold surface for biospecific interaction analysis in surface plasmon resonance sensors. Anal Biochem 1991; 198:268 - 77; http://dx.doi.org/10.1016/0003-2697(91)90424-R; PMID: 1724720