Abstract
The PIF1 helicase family performs many cellular functions. To better understand the functions of the human PIF1 helicase, we characterized the biochemical properties of its ATPase. PIF1 is very sensitive to temperature, whereas it is not affected by pH, and the ATPase activity of human PIF1 is dependent on the divalent cations Mg2+ and Mn2+ but not Ca2+ and Zn2+. Inhibition was observed when single-stranded DNA was coated with RPA or SSB. Moreover, the ATPase activity of PIF1 proportionally decreased with decreasing oligonucleotide length due to a decreased binding ability. A minimum of 10 oligonucleotide bases are required for PIF1 binding and the hydrolysis of ATP. The analysis of the biochemical properties of PIF1 together with numerous genetic observations should aid in the understanding of its cellular functions.
Keywords: :
Introduction
Helicases play important roles in nearly all aspects of DNA metabolism, acting as critical regulators of genomic stability. Thus, mutations in several DNA helicases of the RecQ family are associated with human diseases, such as Werner, Bloom and Rothmund-Thomson syndromes.Citation1-Citation3
The PIF1 subfamily of 5′ to 3′ DNA helicasesCitation4,Citation5 is conserved from yeast to human.Citation6 In Saccharomyces cerevisiae (Sc), ScPif1 was first isolated due to its involvement in recombination and maintenance of mitochondrial DNA.Citation7 Pif1 helicase cooperates with base excision repair to protect against spontaneous oxidative mtDNA damage, and pif1 mutant cells are sensitive to ethidium bromide-induced damage with fragmentation of mtDNA.Citation8-Citation11 Pif1p possesses both nuclear and mitochondrial targeting signals.Citation12 The nuclear ScPif1p performs multiple functions, including the catalytic inhibition of telomerase activity.Citation13 The overexpression of ScPif1p results in shorter telomeres, whereas mutation of ScPif1p results in longer telomeres.Citation12,Citation14 ScPif1p also inhibits replication fork progression in rDNA,Citation15 plays roles in Okazaki fragment procession,Citation16 and participates in resolving G-quadruplex complexes.Citation17
Schizosaccharomyces pombe contains a PIF1 homolog, pfh1+,Citation18 that encodes Pfh1p, which exists as multiple isoforms that localize to either nuclei or mitochondria and is essential for the maintenance of nuclear and mitochondrial DNA.Citation19 Pfh1p is required for the completion of DNA replication and for the proper response to DNA-damaging agents.Citation18 Pfh1p also participates in lagging strand DNA maturation and DNA replication through hard-to-replicate sites.Citation20,Citation21 Due to difficulties in the purification of full-length human PIF1, the first study of hPIF1 was performed on an N-terminally truncated PIF1.Citation22-Citation24 Subsequently, we reported the first successful purification of full-length human PIF1 protein and documented novel functions of the N-terminal (PINT) domain.Citation25 Here, we characterized the biochemical properties of the ATPase activity of full-length human PIF1. Biochemical studies of the human PIF1 protein should aid in understanding its physiological function.
Results
Expression of human PIF1 in human tissue
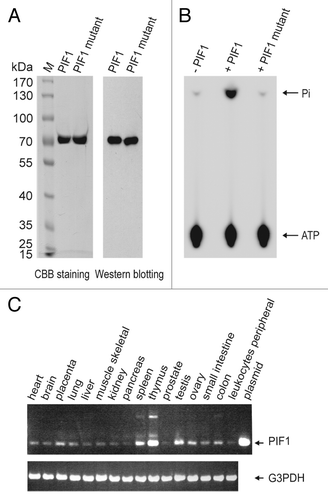
To determine the tissue-specific distribution of PIF1 mRNA, we performed RT-PCR using commercial mul-titissue cDNA panels (). The expression of PIF1 appeared low among 16 tested human tissues, demonstrating relatively higher levels in thymus, spleen and testis and the lowest level in prostate. Of the same tested tissues, our results were consistent with previously reported results.Citation26
Figure 1. Characterization of PIF1. (A) SDS-PAGE of the purified PIF1 and PIF1K234A mutant. Ten micro grams of purified PIF1 and PIF1K234A mutant were loaded on 8% polyacrylamide gel followed by Coomassie brilliant blue R-250 staining and western blotting with an anti-PIF1 antibody. (B) ATPase activity of human PIF1 protein and PIF1K234A mutant. ATPase activities were assayed under standard reaction conditions using M13 mp18 ssDNA (3.8 μM in nucleotides) and without PIF1, with PIF1 (140 nM) or PIF1K234A mutant (140 nM) at 30 °C for 10 min. (C) Expression of PIF1. The expression of PIF1 mRNA was analyzed using RT-PCR and cDNAs from various human tissues. The expression of G3PDH mRNA was independently assayed as a control.
Characterization of ATPase activity
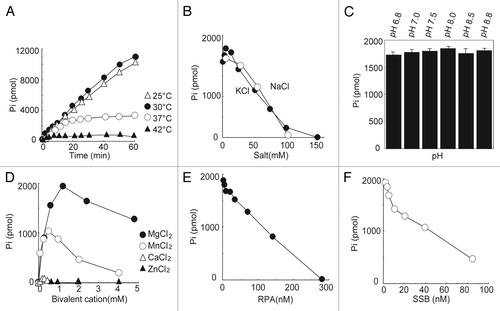
DNA helicases are enzymes that unwind DNA duplexes with the energy released from the hydrolysis of ATP. These enzymes possess ATPase activity, the characterization of which provides important information on their functions as DNA helicases. To determine the biochemical properties of full-length human PIF1, PIF1 containing an N-terminal 6x histidine tag was purified using a previously established procedure (), and the ATPase activity was measured in standard reaction mixtures (Materials and Methods) containing 7.5 μM M13 mp18 ssDNA and 9 nM PIF1 protein. As shown in , PIF1 is very sensitive to heat. At 25 °C and 30 °C, a similar level of activity was noted, and the amount of products proportionally increased with time within 60 min, which resulted in a total release of 11 000 pmol of phosphate. However, with incubation at 37 °C, the increase in the amount of product began to slow after 7 min and ceased after 15 min incubation, which resulted in a total release of 3000 pmol of phosphate. Incubation at 42 °C resulted in the complete inactivation of the ATPase. Salt, in general, inhibited the ATPase activity of PIF1. A slight stimulation was apparent in the presence of a low salt concentration (less than 1.6 mM), but inhibition occurred at higher concentrations (more than 6 mM) of KCl (). NaCl inhibited the ATPase activity of PIF1 (). However, PIF1 did not appear to be sensitive to pH, as no significant difference in the ATPase activity over a wide pH range from 6.8 to 8.8 could be detected using the Student t test for the comparison with the ATPase activity measured at pH 7.5 (). The ATPase activity was dependent on divalent metals, and Mg2+ (1.2 mM) stimulated the ATPase activity to the greatest extent. Mn2+ (0.5 mM), and Ca2+ (0.2 mM) at their optimal concentrations supported 55% and 4% of the Mg2+-supported ATPase activity, whereas 1.2 mM Mn2+ only supported 35% of the Mg2+-supported ATPase activity, and 1.2 mM Ca2+ did not support the ATPase activity. However, Zn2+ did not affect the ATPase activity of PIF1. Because in vivo, single-stranded DNA is typically coated with single-stranded DNA binding proteins, such as RPA in eukaryotes and SSB in prokaryotes, elucidation of whether PIF1 demonstrates ATPase activity in the presence of RPA or SSB is of interest with respect to its cellular functions. Using the determined optimal assay conditions, both RPA and SSB inhibited the ATPase activity of PIF1 over a wide concentration range from 2.0 nM to 280 nM of RPA and from 1.0 nM to 85 nM of SSB, as shown in . Previously we calculated the turnover rate of ATP hydrolysis to be approximately 1000 min−1, which is equivalent to those for ScPif1 and the fission yeast Pfh1.Citation5,Citation20,Citation25 An ATPase-dead mutant, PIF1 K234A, in which the lysine in the ATP-binding domain was mutated to an alanine, was also purified by the same purified procedures (), and its ATPase activity was determined. Under standard reaction conditions, the ATPase activity was below the background level ().
Figure 2. Determination of the biochemical properties of the ATPase activity of human PIF1. (A) Effect of temperature on the stimulation of PIF1 ATPase activity. ATPase activities were assayed under standard reaction conditions using M13 mp18 ssDNA (3.8 μM in nucleotides) and PIF1 (9.0 nM) at 25 °C, 30 °C, 37 °C, and 42 °C for the indicated times. (B) Effects of salt on the stimulation of PIF1 ATPase activity. Assays were conducted under standard reaction conditions using M13 mp18 ssDNA (3.8 μM in nucleotides), PIF1 (9.0 nM) and increasing concentrations of NaCl or KCl at 30 °C for 10 min. (C) Effects of pH on the stimulation of PIF1 ATPase activity. Assays were conducted under standard reaction conditions using M13 mp18 ssDNA (3.8 μM in nucleotides) and PIF1 (9.0 nM) at different pH values and 30 °C for 10 min. The data represent the means ± SD from 3 independent experiments. (D) Effects of divalent ions on the stimulation of PIF1 ATPase activity. Assays were measured under standard reaction conditions using M13 mp18 ssDNA (3.8 μM in nucleotides), PIF1 (9.0 nM) and different divalent ions at 30 °C for 10 min. (E) Effects of RPA on the stimulation of PIF1 ATPase activity. M13 mp18 ssDNA (3.8 μM in nucleotides) was incubated with an increasing concentration of RPA on ice for 10 min under standard reaction conditions. PIF1 (9.0 nM) was introduced at 30 °C for 10 min. (F) Effects of SSB on the stimulation of PIF1 ATPase activity. M13 mp18 ssDNA (3.8 μM in nucleotides) was incubated with an increasing concentration of SSB on ice for 10 min under standard reaction conditions. PIF1 (9.0 nM) was then introduced and incubation continued at 30 °C for 10 min. The data represent 3 independent experiments, and the errors were less than 10%.
Effect of the length of single-stranded DNA on the ATPase activity
Previously, we demonstrated that single-stranded DNA optimally stimulates the ATPase activity of PIF1.Citation25 To determine the single-stranded DNA length requirements for the ATPase activity, we compared the ATPase activity stimulated by six different oligonucleotides, which ranged from 60 to 10 nucleotides in length that were composed of two bases (CT) to ensure the absence of secondary structures. The results revealed a positive correlation between the ATPase activity and oligonucleotide length (). Decreasing the oligonucleotide length from 60 to 30 nucleotides decreased the ATPase activity 15% with each 10-nucleotide increment, whereas a 20-mer oligonucleotide only supported 50% of the ATPase activity that was stimulated by a 30-mer oligonucleotide. The stimulation of the ATPase activity was minimal using a 10-mer oligonucleotide.
Figure 3. Determination of the effect of different oligonucleotide lengths on PIF1 ATPase activity. (A) Effects of different oligonucleotide lengths on PIF1 ATPase activity. ATPase activity was assayed under standard reaction conditions using oligonucleotides (3.8 μM in nucleotides) that ranged in length from 60 to 10 nucleotides and PIF1 (4.5 nM) at 30 °C for 10 min. (B) Titration of oligonucleotides for the stimulation of PIF1 ATPase activity. ATPase activity was measured under the standard reaction conditions at 30 °C for 10 min. The protein concentration was 4.5 nM. The data represent 3 independent experiments, and the errors were less than 10%.
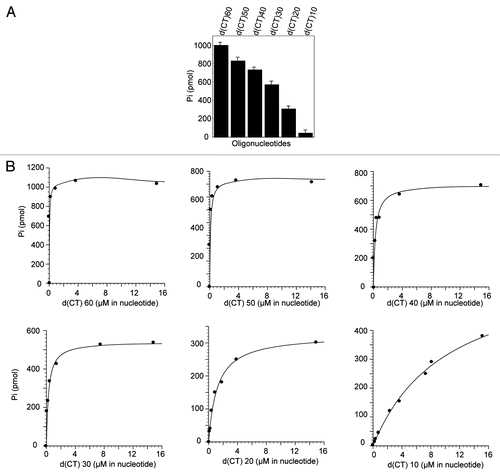
Because these experiments were performed with nearly saturating concentrations of oligonucleotides, the results do not necessarily reflect the affinities of the respective ssDNAs. We therefore determined a kinetic parameter, Keff, that was calculated from ssDNA-titration experiments () using a hyperbolic curve-fitting program (). The results revealed that decreasing the length from 60 to 30 nucleotides increased the Keff values 2-fold with each 10-nucleotide increment. The Keff value for a 20-mer oligonucleotide was 4-fold higher than that for a 30-mer oligonucleotide, which is consistent with the sudden decrease in ATPase activity. The Keff value for a 10-mer oligonucleotide was found to be 7-fold higher than that for a 20-mer oligonucleotide, which explains its failure to stimulate ATPase activity.
Table 1. Kinetic parameter of the ATPase activity, Keff, and apparent Kd of PIF1 binding to various ssDNAs.
Effects of the length of single-stranded DNA on ssDNA binding ability
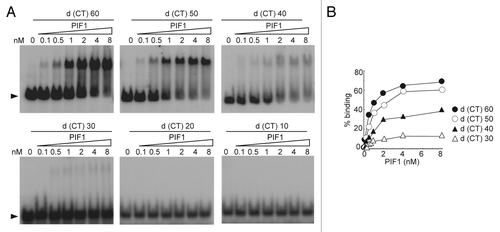
We previously established a method to determine the specific DNA binding activity of PIF1 and determined the ssDNA binding activity of PIF1.Citation25 The decrease in ATPase activity for shorter oligonucleotides might thus be attributable to reduced binding. To measure the ability to bind to different lengths of ssDNA, we performed electrophoretic mobility shift assays (EMSA) using six different oligonucleotides, which ranged from 60 to 10 nucleotides in length that were composed of two bases (CT) to ensure the absence of secondary structures, as in the determination of the optimal ATPase assay conditions. The products were loaded on gels as described in the Materials and Methods section. For the oligonucleotides that contained 60 to 30 nucleotides, the titration of the ssDNA with PIF1 resulted in the formation of PIF1-DNA complexes, which increased with increasing PIF1 concentration (). PIF1-DNA complexes were not detected for oligonucleotides containing 20 or 10 nucleotides. The apparent Kd values, which are approximately equal to the protein concentration at which half of the free DNA becomes bound,Citation27 are listed in . The apparent Kd values for d(CT)60 and d(CT)50 were determined to be approximately 1.29 nM and 2.7 nM, which are in good agreement with their Keff values, 0.05 μM and 0.11 μM (in nucleotides) (), after correction for the oligonucleotide concentration, which results in 0.83 nM and 2.2 nM, respectively. The apparent Kd value for d(CT)40 was estimated to be approximately 15.55 nM, which is slightly higher than the Keff value of 7 nM, which represents the correction of 0.279 μM for the concentration of a 40-mer oligonucleotide. A Kd for d(CT)30 could not be determined due to limited binding.
Figure 4. EMSA of PIF1 binding to oligonucleotides of different lengths. (A) The binding of PIF1 to oligonucleotides of different lengths. 5′-32P-labeled oligonucleotides of different lengths were incubated with the indicated concentrations of PIF1. Arrowheads show the positions of free DNA. (B) Graphic illustration of the quantified data shown for A. The data represent 3 independent experiments, and the errors were less than 10%.
Discussion
The present study of the ATPase activity of the human PIF1 helicase revealed a clear correlation with ssDNA binding ability. The ATPase activity of helicases is necessary to provide the energy to enable the translocation of these enzymes along ssDNA and unwind DNA duplexes. Characterization of the ATPase activity provides overall information on the helicase functions of these enzymes. As shown in , human PIF1 is very sensitive to temperature, which may partially account for the difficulty encountered in its purification. In contrast, the protein is not sensitive to pH, demonstrating a very broad optimal pH. KCl and NaCl, in general, inhibit the ATPase activity of PIF1, which is consistent with previous reports of the fission yeast homolog, Pfh1, and the N-terminally truncated PIF1,Citation18,Citation19 whereas divalent cations exert a stimulatory effect. In the present study, Mg2+ was found to provide the greatest stimulation followed by Mn2+, whereas Ca2+ and Zn2+ demonstrated only weak effects. In vivo, single-stranded DNA is typically coated with RPA in eukaryotes and SSB in prokaryotes,Citation25,Citation28 and both RPA and SSB can inhibit the ATPase activity of PIF1, which suggest that the functions of PIF1 might be regulated by upstream proteins in vivo.
We previously reported that ssDNA binds to PIF1 and greatly stimulates its ATPase activity.Citation25 In the present study, we demonstrated that the ATPase activity decreases proportionally with a decreasing length of ssDNA. The decreased activity could clearly result from the reduced binding of PIF1 to shorter nucleotides. The apparent Kd values for d(CT)60 and d(CT)50 of 1.29 nM and 2.7 nM, respectively, were in good agreement with the Keff values of 0.83 nM and 2.2 nM, respectively, that are expressed in terms of the oligonucleotide concentration. We suggest that the difficulty in binding shorter oligonucleotides directly leads to a decreased PIF1ATPase activity. Because PIF1 does not sufficiently bind short oligonucleotides that are 10 nucleotides in length, the oligonucleotide d(CT)10 did not appreciably stimulate the ATPase activity of PIF1. Notably, the apparent Kd value for d(CT)30 was too high to be determined, and d(CT)20-PIF1 complexes could not be detected, although d(CT)30 and d(CT)20 were able to stimulate PIF1 ATPase activity to an appreciable extent. This discrepancy might be explained by the insufficient sensitivity of EMSA to detect short oligonucleotide binding to PIF1. It should also be noted that the apparent Kd value for d(CT)40 of 15.55 nM is slightly higher than its Keff value of 7 nM for the ATPase activity.
The PIF1 helicase subfamily appears to perform many cellular functions, ranging from nuclear DNA replication,Citation15 telomere length regulation,Citation12-Citation14,Citation26,Citation29,Citation30 mitochondrial genome integrity,Citation7-Citation11,Citation20 DNA repair,Citation8-Citation10 Okazaki fragment processing,Citation16 assisting the replication fork progress through nonnucleosomal protein-DNA complexes.Citation15 Pif1-like helicases are prone to aggregation and are poorly soluble, which hampers their purification, and in vitro studies have, therefore, been very limited. The results of the present analysis of the biochemical properties of PIF1, together with numerous genetic observations, should aid in a better understanding of its cellular functions.
Materials and Methods
Protein purification and antibody production
His-tagged human PIF1K234A point substitution mutation was constructed by QuickChange® Site-Directed Mutagenesis Kit (Stratagene). His-tagged full-length PIF1 and PIF1K234A were purified from overexpression in E. coli BL21 (DE3) cells. The detailed purification procedures have been previously described.Citation25 RPA was purified as previously describedCitation28 and SSB was purchased from Stratagene (Stratagene). Polyclonal antibodies against PIF1 was generated as previously described.Citation25
Multi-tissue RT-PCR
Human MTC Panel I (#636742) and Panel II (#636743) were purchased from Clontech (Clontech), and the forward primer, CACAGATTTG AGGCTATGGA C, and reverse primer, TCAGAGGATT GGGTCCATGT T, were used to amplify PIF1 using RT-PCR. The primers for G3PDH were provided by the manufacturer. Cycling parameters for the analysis of PIF1 were as follows: 96 °C for 30 s, 55 °C for 30 s, and 72 °C for 2 min. A total of 30 cycles of PCR were performed to amplify PIF1 and G3PDH.
ATPase assay
ATPase activity was measured in 50 mM TRIS-HCl (pH 8.0), 2 mM DTT, 1.2 mM MgCl2, 0.25 mg/ml BSA, 2 mM [γ-32P]ATP, the indicated amount of DNA, and the indicated amount of protein sample diluted with buffer D (50 mM TRIS-HCl, pH 8.0, 1 M NaCl, 2 mM DTT, 10% glycerol, and 0.1 mg/ml BSA). After incubation at 30 °C for 10 min, the reaction was stopped with 4 μl of 20 mM EDTA (pH 8.0). Two μl of aliquot was spotted onto a polyethyleneimine-cellulose plate (Merck) and developed in 0.3 M LiCl/0.9 M formic acid. The products were analyzed as previously described.Citation25
DNA binding assays
End-labeled oligonucleotides were labeled with polynucleotide kinase (New England BioLabs) and [γ-32P] ATP. DNA binding assays were performed using a modified method previously described.Citation31 The reactions were performed under ATPase assay conditions using 25 pM end -labeled oligonucleotides and indicated amount of PIF1 protein. Samples were incubated on ice for 10 min and loaded on pre-run 5% polyacrylamide gels (79:1 acrylamide/bisacrylamide). The reaction products were analyzed as previously described.Citation25
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We are grateful to Yuji Masuda for helpful discussions and to Fumie Okubo, Kazumi Shimamoto, Xiaodan Liu and Yu Wang for their laboratory assistance. This work was supported by the Chinese National Natural Science Foundation grants 30960093, 31160183, and 31270894.
References
- Guo RB, Rigolet P, Zargarian L, Fermandjian S, Xi XG. Structural and functional characterizations reveal the importance of a zinc binding domain in Bloom’s syndrome helicase. Nucleic Acids Res 2005; 33:3109 - 24; http://dx.doi.org/10.1093/nar/gki619; PMID: 15930159
- Hanada K, Hickson ID. Molecular genetics of RecQ helicase disorders. Cell Mol Life Sci 2007; 64:2306 - 22; http://dx.doi.org/10.1007/s00018-007-7121-z; PMID: 17571213
- Macris MA, Krejci L, Bussen W, Shimamoto A, Sung P. Biochemical characterization of the RECQ4 protein, mutated in Rothmund-Thomson syndrome. DNA Repair (Amst) 2006; 5:172 - 80; http://dx.doi.org/10.1016/j.dnarep.2005.09.005; PMID: 16214424
- Boulé JB, Zakian VA. The yeast Pif1p DNA helicase preferentially unwinds RNA DNA substrates. Nucleic Acids Res 2007; 35:5809 - 18; http://dx.doi.org/10.1093/nar/gkm613; PMID: 17720711
- Lahaye A, Leterme S, Foury F. PIF1 DNA helicase from Saccharomyces cerevisiae. Biochemical characterization of the enzyme. J Biol Chem 1993; 268:26155 - 61; PMID: 8253734
- Bessler JB, Torredagger JZ, Zakian VA. The Pif1p subfamily of helicases: region-specific DNA helicases?. Trends Cell Biol 2001; 11:60 - 5; http://dx.doi.org/10.1016/S0962-8924(00)01877-8; PMID: 11166213
- Foury F, Kolodynski J. pif mutation blocks recombination between mitochondrial rho+ and rho- genomes having tandemly arrayed repeat units in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 1983; 80:5345 - 9; http://dx.doi.org/10.1073/pnas.80.17.5345; PMID: 6310571
- Doudican NA, Song B, Shadel GS, Doetsch PW. Oxidative DNA damage causes mitochondrial genomic instability in Saccharomyces cerevisiae. Mol Cell Biol 2005; 25:5196 - 204; http://dx.doi.org/10.1128/MCB.25.12.5196-5204.2005; PMID: 15923634
- O’Rourke TW, Doudican NA, Mackereth MD, Doetsch PW, Shadel GS. Mitochondrial dysfunction due to oxidative mitochondrial DNA damage is reduced through cooperative actions of diverse proteins. Mol Cell Biol 2002; 22:4086 - 93; http://dx.doi.org/10.1128/MCB.22.12.4086-4093.2002; PMID: 12024022
- O’Rourke TW, Doudican NA, Zhang H, Eaton JS, Doetsch PW, Shadel GS. Differential involvement of the related DNA helicases Pif1p and Rrm3p in mtDNA point mutagenesis and stability. Gene 2005; 354:86 - 92; http://dx.doi.org/10.1016/j.gene.2005.03.031; PMID: 15907372
- Cheng X, Dunaway S, Ivessa AS. The role of Pif1p, a DNA helicase in Saccharomyces cerevisiae, in maintaining mitochondrial DNA. Mitochondrion 2007; 7:211 - 22; http://dx.doi.org/10.1016/j.mito.2006.11.023; PMID: 17257907
- Zhou J, Monson EK, Teng SC, Schulz VP, Zakian VA. Pif1p helicase, a catalytic inhibitor of telomerase in yeast. Science 2000; 289:771 - 4; http://dx.doi.org/10.1126/science.289.5480.771; PMID: 10926538
- Boulé JB, Vega LR, Zakian VA. The yeast Pif1p helicase removes telomerase from telomeric DNA. Nature 2005; 438:57 - 61; http://dx.doi.org/10.1038/nature04091; PMID: 16121131
- Schulz VP, Zakian VA. The saccharomyces PIF1 DNA helicase inhibits telomere elongation and de novo telomere formation. Cell 1994; 76:145 - 55; http://dx.doi.org/10.1016/0092-8674(94)90179-1; PMID: 8287473
- Ivessa AS, Zhou JQ, Zakian VA. The Saccharomyces Pif1p DNA helicase and the highly related Rrm3p have opposite effects on replication fork progression in ribosomal DNA. Cell 2000; 100:479 - 89; http://dx.doi.org/10.1016/S0092-8674(00)80683-2; PMID: 10693764
- Budd ME, Reis CC, Smith S, Myung K, Campbell JL. Evidence suggesting that Pif1 helicase functions in DNA replication with the Dna2 helicase/nuclease and DNA polymerase delta. Mol Cell Biol 2006; 26:2490 - 500; http://dx.doi.org/10.1128/MCB.26.7.2490-2500.2006; PMID: 16537895
- Ribeyre C, Lopes J, Boulé JB, Piazza A, Guédin A, Zakian VA, Mergny JL, Nicolas A. The yeast Pif1 helicase prevents genomic instability caused by G-quadruplex-forming CEB1 sequences in vivo. PLoS Genet 2009; 5:e1000475; http://dx.doi.org/10.1371/journal.pgen.1000475; PMID: 19424434
- Tanaka H, Ryu GH, Seo YS, Tanaka K, Okayama H, MacNeill SA, Yuasa Y. The fission yeast pfh1(+) gene encodes an essential 5′ to 3′ DNA helicase required for the completion of S-phase. Nucleic Acids Res 2002; 30:4728 - 39; http://dx.doi.org/10.1093/nar/gkf590; PMID: 12409464
- Pinter SF, Aubert SD, Zakian VA. The Schizosaccharomyces pombe Pfh1p DNA helicase is essential for the maintenance of nuclear and mitochondrial DNA. Mol Cell Biol 2008; 28:6594 - 608; http://dx.doi.org/10.1128/MCB.00191-08; PMID: 18725402
- Ryu GH, Tanaka H, Kim DH, Kim JH, Bae SH, Kwon YN, Rhee JS, MacNeill SA, Seo YS. Genetic and biochemical analyses of Pfh1 DNA helicase function in fission yeast. Nucleic Acids Res 2004; 32:4205 - 16; http://dx.doi.org/10.1093/nar/gkh720; PMID: 15302919
- Sabouri N, McDonald KR, Webb CJ, Cristea IM, Zakian VA. DNA replication through hard-to-replicate sites, including both highly transcribed RNA Pol II and Pol III genes, requires the S. pombe Pfh1 helicase. Genes Dev 2012; 26:581 - 93; http://dx.doi.org/10.1101/gad.184697.111; PMID: 22426534
- Futami K, Shimamoto A, Furuichi Y. Mitochondrial and nuclear localization of human Pif1 helicase. Biol Pharm Bull 2007; 30:1685 - 92; http://dx.doi.org/10.1248/bpb.30.1685; PMID: 17827721
- Huang Y, Zhang DH, Zhou JQ. Characterization of ATPase activity of recombinant human Pif1. Acta Biochim Biophys Sin (Shanghai) 2006; 38:335 - 41; http://dx.doi.org/10.1111/j.1745-7270.2006.00165.x; PMID: 16680374
- Zhang DH, Zhou B, Huang Y, Xu LX, Zhou JQ. The human Pif1 helicase, a potential Escherichia coli RecD homologue, inhibits telomerase activity. Nucleic Acids Res 2006; 34:1393 - 404; http://dx.doi.org/10.1093/nar/gkl029; PMID: 16522649
- Gu Y, Masuda Y, Kamiya K. Biochemical analysis of human PIF1 helicase and functions of its N-terminal domain. Nucleic Acids Res 2008; 36:6295 - 308; http://dx.doi.org/10.1093/nar/gkn609; PMID: 18835853
- Mateyak MK, Zakian VA. Human PIF helicase is cell cycle regulated and associates with telomerase. Cell Cycle 2006; 5:2796 - 804; http://dx.doi.org/10.4161/cc.5.23.3524; PMID: 17172855
- Carey J. Gel retardation. Methods Enzymol 1991; 208:103 - 17; http://dx.doi.org/10.1016/0076-6879(91)08010-F; PMID: 1779832
- Masuda Y, Suzuki M, Piao J, Gu Y, Tsurimoto T, Kamiya K. Dynamics of human replication factors in the elongation phase of DNA replication. Nucleic Acids Res 2007; 35:6904 - 16; http://dx.doi.org/10.1093/nar/gkm822; PMID: 17932049
- Pennaneach V, Putnam CD, Kolodner RD. Chromosome healing by de novo telomere addition in Saccharomyces cerevisiae. Mol Microbiol 2006; 59:1357 - 68; http://dx.doi.org/10.1111/j.1365-2958.2006.05026.x; PMID: 16468981
- Mangahas JL, Alexander MK, Sandell LL, Zakian VA. Repair of chromosome ends after telomere loss in Saccharomyces. Mol Biol Cell 2001; 12:4078 - 89; http://dx.doi.org/10.1091/mbc.12.12.4078; PMID: 11739802
- Masuda Y, Takahashi M, Tsunekuni N, Minami T, Sumii M, Miyagawa K, Kamiya K. Deoxycytidyl transferase activity of the human REV1 protein is closely associated with the conserved polymerase domain. J Biol Chem 2001; 276:15051 - 8; http://dx.doi.org/10.1074/jbc.M008082200; PMID: 11278384