 ?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
A sample of purified Syrian hamster PrP27–30 prion fibers was analyzed by synchrotron small-angle X-ray scattering (SAXS). The SAXS pattern obtained was fitted to a model based on infinitely long cylinders with a log-normal intensity distribution, a hard-sphere structure factor and a general Porod term for larger aggregates. The diameter calculated for the cylinders determined from the fit was 11.0 ± 0.2 nm. This measurement offers an estimation of the diameter of PrPSc fibers in suspension, i.e., free of errors derived from estimations based on 2D projections in transmission electron microscopy images, subjected to further possible distortions from the negative stain. This diameter, which corresponds to a maximum diameter of approximately 5.5 nm for each of the two intertwined protofilaments making up the fibers, rules out the possibility that PrPSc conforms to a stack of in-register, single-rung flat PrPSc monomers; rather, PrPSc subunits must necessarily coil, most likely several times, into themselves.
Introduction
Understanding the structure of PrPSc is still one of the major challenges in prion research. The mechanism by which prions propagate, as well as the strain and transmission barrier phenomena, will not be understood until a basic knowledge of the structure of PrPSc is available. There is ample consensus that prion transmission is structurally enciphered.Citation1,Citation2 In fact, it is very likely that an immediate understanding of the essential mechanisms for prion replication and propagation will derive from contemplating the structure of PrPSc, just as the mechanism of inheritance became obvious once the DNA structure was solved.Citation3 Unfortunately, the insoluble nature of PrPSc, and the unavailability of recombinant prions until very recentlyCitation4,Citation5—yet to be scaled up to yield sufficient amounts of material useful for structural studies—have impeded the use of high resolution analytical techniques, such as NMR or X-ray crystallography.Citation6 In this context, a number of studies using lower resolution approaches, such as electron microscopy,Citation7-Citation11 fiber X-ray diffraction,Citation12 limited proteolysis,Citation1,Citation13,Citation14 FTIR,Citation15-Citation17 deuterium-hydrogen exchange followed by mass spectrometry analysisCitation17 or surface chemical derivatizationCitation18 have provided very important constraints and data on the structure of PrPSc. However, this is still insufficient to offer a clear, unequivocal picture of it.
Negative stain transmission electron microscopy (TEM) has yielded important information on PrPSc, such as its ability to form amyloid-type fibers.Citation7-Citation9,Citation12 These fibers appear to consist of two intertwined protofilaments, each one approximately 5 nm wide, so that the apparent diameter of the complete basic double fiber is about 10 nm.Citation12 The amyloid nature of these fibers has been confirmed by X-ray diffraction experiments, which detected the characteristic 4.8 Ǻ meridional reflection.Citation12 In the most complete EM-based analysis of these fibers to date, Sim and Caughey have reported a width of individual RML GPI-anchorless and wild-type PrPSc protofilaments of 3.5 ± 0.6 nm and 3.7 ± 0.6 nm, respectively.Citation9 Wille et al. in turn have reported widths of 4.8 ± 0.8 nm and 5.7 ± 1.1 nm for RML mouse and SC237 Syrian hamster PrP27–30 protofilaments, respectively.Citation12 The width of the individual protofilament, i.e., the stack of individual PrPSc monomers, is certainly a key structural constraint of PrPSc. However, as discussed by Sim and Caughey, determining protofilament diameters from TEM images of negatively stained samples is subject to considerable uncertainty, as it depends on the degree to which the stain pools around the protofilament. Moreover, if only a portion of the protofilaments protrudes from the stain pool, the measured widths will be underestimated. Furthermore, since TEM images represent a 2-dimensional view of a 3-dimensional object, if the filament does not have a regular cross-section, the measurement would be even more uncertain. It is, therefore, quite important to determine the width of PrPSc protofilaments by a method not suffering from these ambiguities.
In this context, small-angle X-ray scattering (SAXS) appears as an attractive option. SAXS provides structural information on macromolecules in solution, based on the Fourier transformation of the electron density contrast of the macromolecules.Citation19,Citation20 Information obtained from SAXS experiments includes the overall shape of the analyzed macromolecule, together with either its approximate radius or its cross sectional diameter, depending on whether the macromolecule is globular or tubular, respectively. The availability of highly luminous synchrotron sources, together with the development of powerful computer-intensive data analysis and fitting algorithms, has allowed for ab initio modeling of protein and fibrillar structures,Citation21,Citation22 placing SAXS at the interface of high resolution techniques such as X-ray crystallography and electron microscopy.Citation23,Citation24 SAXS has been successfully applied to analyze the shape and provide diameter information of tubular structures such as insulin amyloid fibrilsCitation25 and fibrillation of α-synuclein.Citation26 Here, we describe SAXS-based analysis of PrP27–30 fibers prepared from the brains of Syrian hamsters infected with scrapie (SHaPrP27–30).
Results
A highly pure sample of SHaPrP27–30 was obtained as confirmed by SDS-PAGE followed by Coomassie staining (). Virtually no other bands, besides the intense one at 27–30 kDa, corresponding to diglycosylated, N-terminally truncated PrPSc, were detected. A faint smear corresponding to mono- and non-glycosylated PrPSc was visible below such band. A thin band of much lower intensity, corresponding to residual ferritin (as confirmed by proteomic analysis, data not shown) was the only significant impurity present. Immunoassay with the PrP specific antibody 3F4 confirmed the presence of PrP27–30 with predominant diglycosylated and monoglycosylated amino-truncated, PK-resistant PrP ().
Figure 1. (A) Coomassie-stained SDS-PAGE of the SHaPrP27–30 sample subjected to SAXS analysis. (B) After SDS-PAGE, a sample was electroblotted onto a PVDF membrane and blotted with antibody 3F4. (C) Negative stain TEM of the same sample.
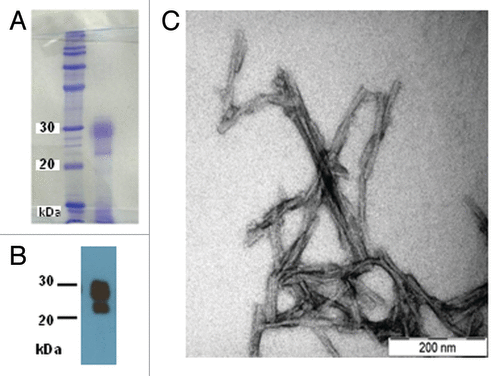
Negative-stain TEM analysis showed abundant double fibers constituted by two intertwined protofilaments. These fibers often packed and/or bundled laterally (). Some additional non-fibrillar material, presumably lipid remnants, was seen at times. These images were very similar to those previously reported in the literature.Citation8,Citation9,Citation12 The estimated width of protofilaments was ∼5 nm, very close to that recently reported by Wille et al.Citation12
The SAXS pattern obtained from this sample, as well as the full pattern fit, are shown in . The model was based on infinitely long cylinders with a log-normal intensity distribution, a hard-sphere structure factor and a general Porod term for larger aggregates of the fiber assembly. shows the fit result assuming the size of 5 nm (derived from TEM of the two protofilaments) and a relative size distribution width of 2.5. The main structural parameters obtained by the fit are the hard-sphere interaction radius (Rhs) and the apparent volume fraction of the hard-spheres (phs). Rhs is equivalent to the distance between the centers of two cylinders touching one another in the sample. It is counterintuitive that we have to introduce in the equation an estimated value for the cylinder diameter (obtained from the TEM images), since what we want to calculate is, precisely, the cylinder diameter (derived from Rhs). However, this apparent paradox is solved if we consider that the contribution of this term to the final result is very small. Thus, if the cylinder form factor is varied with diameters between 2 and 8 nm around the mean value of 5 nm, due to the 1/q dependence of the form factor, the fit results do not change noticeably (). Detailed results are reported in . As the main result, the hard-sphere diameter (2*Rhs) resulted to be 11.0 ± 0.2 nm, which is nearly independent of the initial value estimated from TEM.
Figure 2. SAXS data in log-linear (A) representations and fit results of the infinite cylinder model (R = 5 nm) with hard-sphere structure factor and generalized Porod slope of SHaPrP27–30 (1 mg/mL). In the log-log representation of the SAXS data. (B) fit results varying the radius of the cylinders between 2 and 8 nm are shown together with the generalized Porod contribution (black). The arrow in both graphs indicates the hump of the structure factor corresponding to 11.0 ± 0.2 nm.
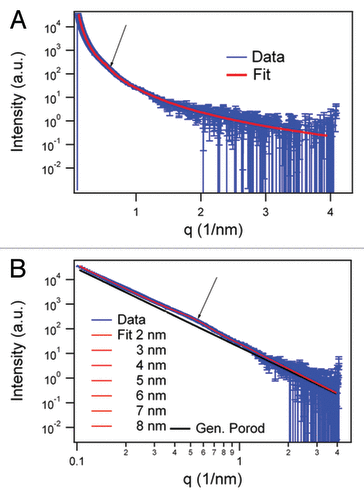
Table 1. Overview of the SAXS fit results using the cylindrical model with hard sphere structure factor and generalized Porod slope
Discussion
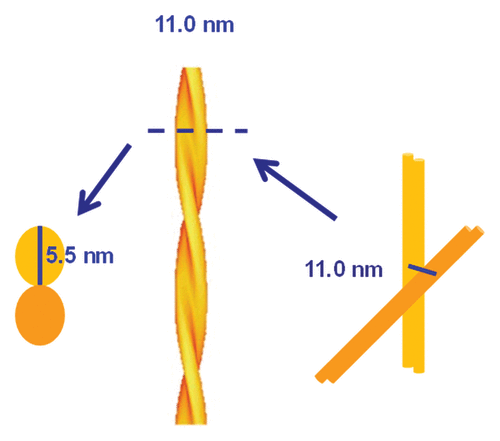
By using SAXS, we obtained from the structure factor a direct measurement of the hard-sphere diameter of SHaPrP27–30 fibers, which is 11.0 ± 0.2 nm. Considering that the hard-sphere diameter corresponds to the diameter of the cylindrical model, this sets a ~5.5 nm constraint for the maximum dimension of the two individual PrPSc protofilaments that, intertwining, constitute the fiber ().
Figure 3. Schematic representation of PrPSc basic fibers based on the information obtained from SAXS and negative stain TEM.
Although the hard-sphere diameter of the 2Rhs can be obtained from SAXS data corresponding to the diameter of fibers, the detailed shape of the cross section was not accessible. Refining the data with an ellipsoidal cross section, which would be a better model for the fiber assembly, gave no reliable results. However, the fits estimated the correct order of magnitude for the eccentricity and dimension (data not shown).
A width of 5.5 nm at its maximum span places constraints, in turn, on the possible architectures of the staked PrPSc monomers that constitute the PrPSc protofilament. Since SHaPrP27–30 contains ~141 amino acid residues, spanning from residue Gly90 to the C-terminal residue Ser231,Citation1,Citation10,Citation13 an extended, in-register stack architecture similar to those proposed for Ure2p and Sup35Citation27,Citation28 can be ruled out, as it is impossible for 141 residues to fit in a flat disk, 5.5 nm wide and 0.5 nm tall (the approximate height of a stackable β-strand). This means that PrPSc must necessarily coil into itself, like the Het-s prion domain,Citation29,Citation30 thus supporting the notion that PrPSc is probably a β-solenoid, as proposed by Wille and Govaerts.Citation10-Citation12 Of note, this proposal has its origin in geometric considerations that assessed different possible structures fitting width constraints derived from 2D crystals seen from the z axis.Citation10,Citation11
The molecular mass of SHaPrP27–30 is 27–30 kDa. Taking 28.5 kDa as a mean value, considering 0.81 Da/ǺCitation3 as the average protein density,Citation31 simplistically assuming a similar density for its carbohydrate component and a basic cylinder shape, the length occupied by a PrPSc molecule in the PrPSc protofilament would be (28.500/0.81)/(27.5)2π where 27.5 Ǻ is the radius of the cylinder of a protofilament derived from our SAXS measurements. This results in a length of 14.5 Ǻ, i.e., equivalent to approximately 3 rungs of β-helical core. Obviously, this highly speculative calculation ignores a number of key uncertainties, such as the exact density of the carbohydrate and GPI, and the fact that the shape, and therefore the volume of the protofilament, will certainly not be those of a perfect cylinder. Still, this calculation illustrates that 2 rungs, as in the case of the Het-s prion domain, is most probably an insufficient height, and 3–4 is a much more realistic figure. This would reasonably agree with fiber X-ray diffraction data reported by Wille et al., strongly suggestive of a 4-rung β-solenoid architecture for PrP27–30 monomers in the amyloid stack.Citation12
Cobb et al. have proposed a model for recombinant PrP amyloid fibers derived from EPR measurements in which each PrP stacks in-register with the upper and lower ones.Citation32 In this model, only the stretch spanning from position 164 to 231 forms the β-strand-rich stackable core; this stretch bends into itself forming a hairpin-like structure similar to that of other amyloids, such as Aβ.Citation33,Citation34 The width of such hairpin-like structure is ∼12 nm, which agrees with the apparent width of these fibers in negative stain TEM images. This further illustrates the absolute requirement of self-coiling of the PrP (90–231) polypeptide chain to form the PrPSc structure, whose β-strand-rich stackable core comprises roughly twice as many residues, to be fitted in approximately half the width.
In summary, for the first time to our knowledge, we estimated the diameter of PrPSc fibers in suspension, i.e., free of possible errors and interferences arising from 2D projections and staining. The approximate diameter thus obtained rules out the possibility that the architecture of PrPSc conforms to a stack of in-register, single-rung flat PrPSc monomers; rather, PrPSc subunits must necessarily coil, most likely several times, into themselves.
Materials and Methods
Isolation of SHaPrP27–30
Syrian hamsters were inoculated intracranially with 10% 263K scrapie-infected hamster brain homogenate. After approximately 90 d, at the terminal stage of the clinical phase, animals were killed and their brain removed and stored at -80 °C until further use. SHaPrP27–30 was isolated from brains according to Raymond et al.Citation35 with a minor modification to better remove ferritin from the preparation. Namely, 10 brains were cut into small pieces and rinsed with ice-cold PBS twice to remove remnants of blood. Pieces were transferred to 85 mL of a 320 mM sucrose solution and homogenized using a 40 mL dounce grinder (Wheaton), with 10 strokes of the loose pestle followed by 10 strokes of the tight pestle. The resulting suspension was transferred to 4 polycarbonate tubes (Beckman Coulter) and centrifuged for 30 min at 35000 rpm in a Ti 70 rotor at 20 °C. The translucent, pink-reddish supernatants, containing most ferritin, were discarded, and the whitish, fluffy, membrane-rich pellets saved, leaving ~3 mL of supernatant so as to not to disturb the pellet. Twenty mL of 320 mM sucrose was added to each tube and pellets homogenized with a plastic Pasteur pipette followed by vortexing. Samples were spun again as described. The much lighter pink supernatants were discarded, and the more compact membrane-rich pellets containing PrPSc were resuspended in 20 mL of 20 mM Tris, pH 8.5, containing 100 mM NaCl and 10% sarkosyl. Homogenates were pooled and further homogenized using a dounce grinder as described above. The homogenate was let stand at room temperature for 30 min and transferred to 4 polycarbonate tubes. PrPSc isolation proceeded from this point on exactly as described by Raymond et al., including PK treatment, except that a 70 Ti rotor was used. The final SHaPrP27–30 pellet was suspended in 350 µL of deionized water. The sample was characterized by SDS-PAGE in a 15% gel followed by Coomassie staining. In parallel, the gel was electroblotted and probed with mAB 3F4.
Negative stain TEM
Ten µL of SHaPrP27–30 suspension were deposited on a carbon-coated copper grid (Ted Pella, Redding), allowed to adsorb for 5 min, washed twice with deionized water and stained for 5 min with a freshly prepared, filtered, 5% solution of uranyl acetate. Grids were gently blotted with filter paper and allowed to dry overnight. Samples were analyzed by TEM on a Philips CM-12 electron microscope at100 kV.
SAXS analysis
The SHaPrP27–30 suspension was placed in a glass capillary tube with 1.5 mm diameter (Hilgenberg GmbH) and analyzed at the Austro SAXS beamline (5.2L) of the electron storage ring ELETTRA.Citation36 The suspension had an approximate concentration of 1 mg/mL. A Mar300 imageplate reader (MarResearch GmbH) was placed at a distance of 1639.4 mm for recording the X-ray images, whose scattering vector scale was calibrated with Ag behenate (d-spacing: 5.8376 nm).Citation37 The data were corrected for transmission and fluctuations of the primary beam, and background subtracted using a buffer solution as reference.
SAXS data analysis and fitting
The measured X-ray images were integrated with Fit2DCitation38 and the one dimensional data analyzed with software package IgorPro (Wavemetrics). The model applied was an interacting infinite long cylinder model with a log-normal intensity size distribution for the cylinder radii. For simplicity, a hard-sphere modelCitation39 was used to describe the structure factor with the local monodisperse assumption. Additionally, to take into account the hierarchical architecture of the fiber assembly, which is beyond the q-resolution of the SAXS experiment, a general Porod term was added. The full fitting function used was:
where the constant A stands for the intensity fit parameter, R and ΔR for the radius and standard deviation of the cylinder radius, J1 for the 1st order Bessel function, Shardsphere(q,phs,Rhs) for the hard sphere structure factor (with volume fraction p and hard sphere radius Rhs) and cp, p for parameters of the generalized Porod function. B is an optional background.
| Abbreviations: | ||
| GPI | = | glycosyl-phosphatidylinositol |
| PrP | = | prion protein |
| PrPSc | = | prion protein scrapie |
| SAXS | = | small angle X-ray scattering |
| TEM | = | transmission electron microscopy |
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This study was supported by grants FOOD-CT-2006-023183 (STRAINBARRIER) and FP7 222887 (PRIORITY) from the European Union, and BFU2006-04588/BMC from the Spanish Ministry of Science and Education.
References
- Prusiner SB. Prions. Proc Natl Acad Sci U S A 1998; 95:13363 - 83; http://dx.doi.org/10.1073/pnas.95.23.13363; PMID: 9811807
- Safar J, Wille H, Itri V, Groth D, Serban H, Torchia M, Cohen FE, Prusiner SB. Eight prion strains have PrP(Sc) molecules with different conformations. Nat Med 1998; 4:1157 - 65; http://dx.doi.org/10.1038/2654; PMID: 9771749
- Watson JD, Crick FH. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature 1953; 171:737 - 8; http://dx.doi.org/10.1038/171737a0; PMID: 13054692
- Legname G, Baskakov IV, Nguyen HO, Riesner D, Cohen FE, DeArmond SJ, Prusiner SB. Synthetic mammalian prions. Science 2004; 305:673 - 6; http://dx.doi.org/10.1126/science.1100195; PMID: 15286374
- Wang F, Wang X, Yuan CG, Ma J. Generating a prion with bacterially expressed recombinant prion protein. Science 2010; 327:1132 - 5; http://dx.doi.org/10.1126/science.1183748; PMID: 20110469
- Legname G, Giachin G, Benetti F. Structural Studies of Prion Proteins and Prions. In: Rahimi F, Bitan G, eds. Non-fibrillar Amyloidogenic Protein Assemblies-Common Cytotoxins Underlying Degenerative Diseases: Springer, 2012:289-317.
- Merz PA, Somerville RA, Wisniewski HM, Iqbal K. Abnormal fibrils from scrapie-infected brain. Acta Neuropathol 1981; 54:63 - 74; http://dx.doi.org/10.1007/BF00691333; PMID: 7195134
- McKinley MP, Meyer RK, Kenaga L, Rahbar F, Cotter R, Serban A, Prusiner SB. Scrapie prion rod formation in vitro requires both detergent extraction and limited proteolysis. J Virol 1991; 65:1340 - 51; PMID: 1704926
- Sim VL, Caughey B. Ultrastructures and strain comparison of under-glycosylated scrapie prion fibrils. Neurobiol Aging 2009; 30:2031 - 42; http://dx.doi.org/10.1016/j.neurobiolaging.2008.02.016; PMID: 18394757
- Wille H, Michelitsch MD, Guenebaut V, Supattapone S, Serban A, Cohen FE, Agard DA, Prusiner SB. Structural studies of the scrapie prion protein by electron crystallography. Proc Natl Acad Sci U S A 2002; 99:3563 - 8; http://dx.doi.org/10.1073/pnas.052703499; PMID: 11891310
- Govaerts C, Wille H, Prusiner SB, Cohen FE. Evidence for assembly of prions with left-handed beta-helices into trimers. Proc Natl Acad Sci U S A 2004; 101:8342 - 7; http://dx.doi.org/10.1073/pnas.0402254101; PMID: 15155909
- Wille H, Bian W, McDonald M, Kendall A, Colby DW, Bloch L, Ollesch J, Borovinskiy AL, Cohen FE, Prusiner SB, et al. Natural and synthetic prion structure from X-ray fiber diffraction. Proc Natl Acad Sci U S A 2009; 106:16990 - 5; http://dx.doi.org/10.1073/pnas.0909006106; PMID: 19805070
- Sajnani G, Pastrana MA, Dynin I, Onisko B, Requena JR. Scrapie prion protein structural constraints obtained by limited proteolysis and mass spectrometry. J Mol Biol 2008; 382:88 - 98; http://dx.doi.org/10.1016/j.jmb.2008.06.070; PMID: 18621059
- Vázquez-Fernández E, Alonso J, Pastrana MA, Ramos A, Stitz L, Vidal E, Dynin I, Petsch B, Silva CJ, Requena JR. Structural organization of mammalian prions as probed by limited proteolysis. PLoS One 2012; 7:e50111; http://dx.doi.org/10.1371/journal.pone.0050111; PMID: 23185550
- Caughey B, Raymond GJ, Bessen RA. Strain-dependent differences in beta-sheet conformations of abnormal prion protein. J Biol Chem 1998; 273:32230 - 5; http://dx.doi.org/10.1074/jbc.273.48.32230; PMID: 9822701
- Pan KM, Baldwin M, Nguyen J, Gasset M, Serban A, Groth D, Mehlhorn I, Huang Z, Fletterick RJ, Cohen FE, et al. Conversion of alpha-helices into beta-sheets features in the formation of the scrapie prion proteins. Proc Natl Acad Sci U S A 1993; 90:10962 - 6; http://dx.doi.org/10.1073/pnas.90.23.10962; PMID: 7902575
- Smirnovas V, Baron GS, Offerdahl DK, Raymond GJ, Caughey B, Surewicz WK. Structural organization of brain-derived mammalian prions examined by hydrogen-deuterium exchange. Nat Struct Mol Biol 2011; 18:504 - 6; http://dx.doi.org/10.1038/nsmb.2035; PMID: 21441913
- Gong B, Ramos A, Vázquez-Fernández E, Silva CJ, Alonso J, Liu Z, Requena JR. Probing structural differences between PrP(C) and PrP(Sc) by surface nitration and acetylation: evidence of conformational change in the C-terminus. Biochemistry 2011; 50:4963 - 72; http://dx.doi.org/10.1021/bi102073j; PMID: 21526750
- Porod G, Glatter O, Kratky O. Small angle X-ray scattering. In: Glatter O, Kratky O, eds. Academic, London, 1982:17-51.
- Koch MH, Vachette P, Svergun DI. Small-angle scattering: a view on the properties, structures and structural changes of biological macromolecules in solution. Q Rev Biophys 2003; 36:147 - 227; http://dx.doi.org/10.1017/S0033583503003871; PMID: 14686102
- Petoukhov MV, Franke D, Shkumatov AV, Tria G, Kikhney AG, Gajda M, et al. New developments in the ATSAS program package for small-angle scattering data analysis. J Appl Cryst 2012; 45:342 - 50; http://dx.doi.org/10.1107/S0021889812007662
- Petoukhov MV, Svergun DI. Global rigid body modeling of macromolecular complexes against small-angle scattering data. Biophys J 2005; 89:1237 - 50; http://dx.doi.org/10.1529/biophysj.105.064154; PMID: 15923225
- Petoukhov MV, Svergun DI. Applications of small-angle X-ray scattering to biomacromolecular solutions. Int J Biochem Cell Biol 2013; 45:429 - 37; http://dx.doi.org/10.1016/j.biocel.2012.10.017; PMID: 23142499
- Hura GL, Menon AL, Hammel M, Rambo RP, Poole FL 2nd, Tsutakawa SE, Jenney FE Jr., Classen S, Frankel KA, Hopkins RC, et al. Robust, high-throughput solution structural analyses by small angle X-ray scattering (SAXS). Nat Methods 2009; 6:606 - 12; http://dx.doi.org/10.1038/nmeth.1353; PMID: 19620974
- Vestergaard B, Groenning M, Roessle M, Kastrup JS, van de Weert M, Flink JM, Frokjaer S, Gajhede M, Svergun DI. A helical structural nucleus is the primary elongating unit of insulin amyloid fibrils. PLoS Biol 2007; 5:e134; http://dx.doi.org/10.1371/journal.pbio.0050134; PMID: 17472440
- Giehm L, Svergun DI, Otzen DE, Vestergaard B. Low-resolution structure of a vesicle disrupting α-synuclein oligomer that accumulates during fibrillation. Proc Natl Acad Sci U S A 2011; 108:3246 - 51; http://dx.doi.org/10.1073/pnas.1013225108; PMID: 21300904
- Kryndushkin DS, Wickner RB, Tycko R. The core of Ure2p prion fibrils is formed by the N-terminal segment in a parallel cross-β structure: evidence from solid-state NMR. J Mol Biol 2011; 409:263 - 77; http://dx.doi.org/10.1016/j.jmb.2011.03.067; PMID: 21497604
- Shewmaker F, Kryndushkin D, Chen B, Tycko R, Wickner RB. Two prion variants of Sup35p have in-register parallel beta-sheet structures, independent of hydration. Biochemistry 2009; 48:5074 - 82; http://dx.doi.org/10.1021/bi900345q; PMID: 19408895
- Wasmer C, Lange A, Van Melckebeke H, Siemer AB, Riek R, Meier BH. Amyloid fibrils of the HET-s(218-289) prion form a beta solenoid with a triangular hydrophobic core. Science 2008; 319:1523 - 6; http://dx.doi.org/10.1126/science.1151839; PMID: 18339938
- Mizuno N, Baxa U, Steven AC. Structural dependence of HET-s amyloid fibril infectivity assessed by cryoelectron microscopy. Proc Natl Acad Sci U S A 2011; 108:3252 - 7; http://dx.doi.org/10.1073/pnas.1011342108; PMID: 21300906
- Jiang QX, Thrower EC, Chester DW, Ehrlich BE, Sigworth FJ. Three-dimensional structure of the type 1 inositol 1,4,5-trisphosphate receptor at 24 A resolution. EMBO J 2002; 21:3575 - 81; http://dx.doi.org/10.1093/emboj/cdf380; PMID: 12110570
- Cobb NJ, Sönnichsen FD, McHaourab H, Surewicz WK. Molecular architecture of human prion protein amyloid: a parallel, in-register beta-structure. Proc Natl Acad Sci U S A 2007; 104:18946 - 51; http://dx.doi.org/10.1073/pnas.0706522104; PMID: 18025469
- Petkova AT, Ishii Y, Balbach JJ, Antzutkin ON, Leapman RD, Delaglio F, Tycko R. A structural model for Alzheimer’s beta -amyloid fibrils based on experimental constraints from solid state NMR. Proc Natl Acad Sci U S A 2002; 99:16742 - 7; http://dx.doi.org/10.1073/pnas.262663499; PMID: 12481027
- Lührs T, Ritter C, Adrian M, Riek-Loher D, Bohrmann B, Döbeli H, Schubert D, Riek R. 3D structure of Alzheimer’s amyloid-beta(1-42) fibrils. Proc Natl Acad Sci U S A 2005; 102:17342 - 7; http://dx.doi.org/10.1073/pnas.0506723102; PMID: 16293696
- Raymond GJ, Chabry J. Purification of the pathological isoform of prion protein (PrPSc or PrPres) from transmissible spongiform encephalopathy-affected brain tissue. In: Lehmann S, Grassi J, eds. Techniques in Prion Research. Basel: Birkhauser Verlag, 2004:16-24.
- Amenitsch H, Bernstorff S, Kriechbaum M, Lombardo D, Mio H, Rappolt M, et al. Performance and first results of the ELETTRA high-flux beamline for small-angle X-ray scattering. J Appl Cryst 1997; 30:872 - 6; http://dx.doi.org/10.1107/S0021889897001593
- Huang TC, Toraya H, Blanton TN, Wu Y. X-ray powder diffraction analysis of silver behenate, a possible low-angle diffraction standard. J Appl Cryst 1993; 26:180 - 4; http://dx.doi.org/10.1107/S0021889892009762
- Hammersley A. Fit2D V10.3 Reference Manual V4.0. European Synchrotron Radiation Facility, 1997.
- Pedersen JS. Analysis of small-angle scattering data from colloids and polymer solutions: modeling and least-squares fitting. Adv Colloid Interface Sci 1997; 70:171 - 210; http://dx.doi.org/10.1016/S0001-8686(97)00312-6