Abstract
Prions exist as strains, which are thought to reflect PrPSc conformational variants. Prion strains can mutate and it has been proposed that prion mutability depends on an intrinsic heterogeneity of prion populations that would behave as quasispecies. We investigated in vitro prion mutability of 2 strains, by following PrPSc variations of populations serially propagated in PMCA under constant environmental pressure. Each strain was propagated either at low dilution of the seed, i.e., by large population passages, or at limiting dilution, mimicking bottleneck events. In both strains, PrPSc conformational variants were identified only after large population passages, while repeated bottleneck events caused a rapid decline in amplification rates. These findings support the view that mutability is an intrinsic property of prions.
Introduction
According to the protein only hypothesis, prions consist mainly, if not solely, of PrPSc, aggregate conformers of cellular prion protein (PrPC). PrPSc acts as a template and triggers the conversion of PrPC molecules in newly formed PrPSc, leading to prion propagation. This model is supported by protein misfolding cyclic amplification (PMCA),Citation1 a technique that mimic in vitro the autocatalytic PrPC to PrPSc conversion, by seeding healthy brain homogenates as a source of PrPC with minute amounts of prions, and fostering the reaction through multiple cycles of sonication and incubation, which ultimately leads to a vast excess of infectious prions.Citation2
Prions exist as strains that differ in their pathological phenotype and faithfully propagate in the same host, being thus associated with PrPSc having the same amino acid sequence; lacking any evidence of genetic information, it has been proposed that strains represent different PrPSc conformers.Citation3-Citation6 Although strain characteristics are usually maintained on serial passages in the same host, seminal studies have shown that prion strains can mutate upon experimental transmission to a different host, or even when serially passaged into the same host.Citation7 More recently, it has been shown that crossing in vitro the species barrier between mice and hamsters by heterologous PMCA led to the generation of new strains,Citation8 thus mimicking in vitro previous observations. Furthermore, even in absence of sequence mismatches between PrPSc and PrPC, changes in replication environment, such as using RNA-depleted or deglycosilated brain homogenates for prion propagation by PMCA, led to the emergence of variant PrPSc conformers and to strain mutation.Citation9,Citation10
While prion mutation during interspecies transmissions could be explained by the species barrier, which imposes new conformational constraints to PrPSc propagation due to amino acid mismatches with the host PrPC,Citation6,Citation11 it has been less clear how strain mutation occurs during propagation in the same host. Recent advances in this field were made possible by the development of sensitive and strain-selective cell-based assays, such as the cell panel assay,Citation12 allowing to study prion propagation under different selective environments and to differentiate strains based on their cell tropism and sensitivity to various drugs.Citation13 These studies provided strong evidence that prions undergo mutationCitation14 and selective amplificationCitation15 when grown in cell culture, showing that prion populations are intrinsically heterogeneous and exhibit features of Darwinian evolution. Interestingly, in several instances mutated drug-resistant populations positively selected in the presence of drug, reverted upon in vivo passage or by removal of the drug.Citation14,Citation16,Citation17 This led the authors to hypothesize that the behavior of prion populations could be explained in the framework of the quasispecies theory,Citation13 similarly to error prone replication organisms such as RNA viral populations.Citation18 In this scenario, a prion strain would be composed by an ensemble of different conformational molecules that are maintained under host selection; when the host selection changes, the ensemble would shift from one to another dominant conformational molecule. Similar conclusions have been drawn in studies dealing with prions in fungi.Citation19
We have previously shown that bank voles are a sensitive and permissive bioassay for many different prion strains,Citation20-Citation23 and that these features are reflected when using vole brain homogenates as substrate for prion amplification.Citation24 Having set up extremely sensitive and controlled conditions for in vitro amplification of prions by using bank vole PrPC as an ultra-efficient substrate,Citation24 in this study we aimed at investigating the intrinsic mutational ability of prion strains in serial homologous PMCA reactions, in the absence of strong selective constraints such as PrP sequence mismatches, PrPC modifications, RNA-depletion or the addition of drugs. To this aim, we explored the evolutionary potential of 2 prion strains submitted to different passage regimens, i.e., either propagated through large population passages or subjected to repeated bottleneck events.
Results
In preliminary studies aimed at verify the consistency of PMCA reactions, we found that the position of tubes in the PMCA rack was associated with prion replication kinetics, putatively due to an irregular distribution of ultrasound waves during sonication. By reducing the number of tubes in the PMCA rack (Supplemental Material (text); Fig. S1) this effect was considerably reduced. Thus, all experiments were done using only 24 tubes placed in the central portion of the rack and changing the position of each tube at every PMCA round.
Two vole prion strains were selected for the experiments: v586, derived in previous studies from vole PMCA-generated PrPSc and then passaged in vivo,Citation24 characterized by a high replication efficiency () and a BSE-like protease-resistant core of PrPSc (PrPres)Citation24; and SS21, a vole-adapted scrapie strain, with 100 fold less efficient replication efficiency () and a scrapie-like PrPres type.Citation22,Citation25
Figure 1. Experimental design. (A) Serial 10-fold dilutions of v586 and SS21 were subjected to a single round of PMCA, PK digested and analyzed by western blotting with antibody SAF84. The amplification rate was derived from the last positive dilution yielding high PrPres signal, and was ∼107 for v586 and ∼105 for SS21. In the firsts three lanes of each blot were loaded the dilutions 10−2, 10−3, and 10−4 of the inoculum not subjected to PMCA. These values were used to set up our experimental design. (B) PMCA experiments using normal brain homogenates (in blue) as substrate were seeded with brain homogenates from terminally affected voles (in red), and serially propagated through successive PMCA rounds by using two parallel experimental regimens: high PrPSc/PrPC seeding (upper panel), in which prions were continuously passaged by 1:10 dilution; low PrPSc/PrPC seeding (lower panel) which starts from an highly diluted seed and then is propagated by alternating bottleneck passages and large population passages in order to recover the population size.
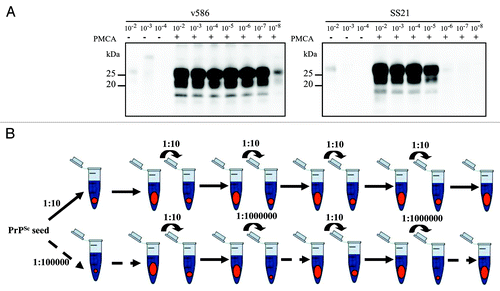
Our aim was to compare the outcome, in terms of PrPres typing, of prion populations replicated for serial 48h PMCA rounds in high or low PrPSc/PrPC ratio, in an attempt to mimic large populations passages (high PrPSc/PrPC) vs. bottleneck events (low PrPSc/PrPC). Unexpectedly, PrPSc was progressively lost in preliminary experiments with v586 continuously propagated at low PrPSc/PrPC (i.e., seeded at 10−7 in serial PMCA), although v586 initially gave high PrPSc yields in similar experiments ( and ref. Citation24). To overcome this effect, bottleneck events were mimicked by alternating 10−7 and 10−2 seeded PMCA rounds (). Five v586 populations were studied with this new set-up, 2 propagated with large population passages (v586/a, v586/b), and 3 subjected to bottleneck events (v586/c, v586/d, v586/e). None of the populations which underwent bottleneck events were lost, although v586/d and v586/e showed a decrease in PrPSc (), suggesting a decrease in replication rate. After 5 PMCA rounds, i.e., a replication factor of ~105 for the populations propagated at high PrPSc/PrPC and ~1020 for those at low PrPSc/PrPC, an upward shift of PrPres was observed only in population v586/a, subjected to high PrPSc/PrPC replication, but not in the three populations which underwent bottleneck events (). This different PrPres pattern emerged after 2 PMCA rounds and was propagated until the fifth round ().
Figure 2. Evolution of PrPSc in v586. (A) western blot analysis of PrPres from v586-derived populations, as indicated. PrPres from different PMCA rounds (roman numbers) were analyzed using antibody SAF84, showing an upward shift of v586/a, and poor PrPres yields in odd rounds of v586/c, v586/d and v586/e. (B) Epitope mapping of PrPres from v586-derived populations analyzed in replica blots with antibodies SAF84 or 12B2, in comparison with v586 and a scrapie control. v586/a showed an intermediate PrPres profile between the inoculum (BSE-like PrPres profile) and the control scrapie. (C) Graph depicting the SAF84/12B2 antibody ratio (y axis, log scale) for samples shown in panel B.
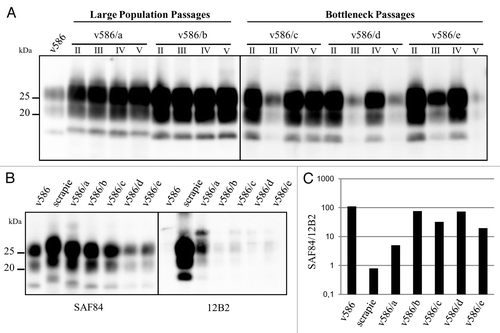
In order confirm to that the slight increase in molecular weight of PrPres observed in v586/a was indeed due to a different PK cleavage site of PrPSc, we used a sensitive discriminative western blot which measures the relative binding of C-terminal and N-terminal anti-PrP mAbs to PrPSc treated with high concentration of PK.Citation26-Citation28 By choosing the N-terminal mAb directed to an epitope near the expected PK cleavage site, the C-terminal/N-terminal ratio reflects the proportion of PrPres molecules having the epitope chosen (N-terminal mAb) compared with the whole population (C-terminal mAb). As v586 is mostly cleaved C-terminally to the epitope of 12B2,Citation24 it was anticipated that the upward shift of v586/a should have been reflected by a sensible increase in 12B2 binding and by a decrease of the ratio between SAF84 (PrP-core antibody) and 12B2. Indeed, v586/b-to-e populations were similar to the original v586 and showed high SAF84/12B2 ratio, while the higher MW of v586/a () was accompanied by a decrease of SAF84/12B2 ratio ().
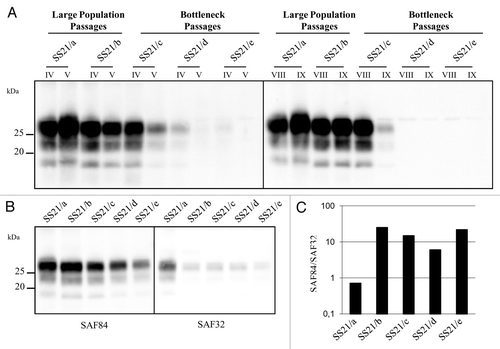
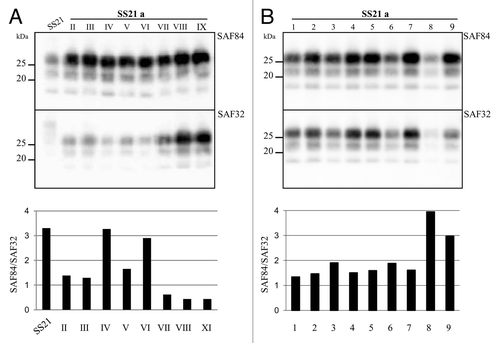
A similar experimental design (high PrPSc/PrPC seeded at 10−2 and low PrPSc/PrPC alternatively seeded at 10−5 and 10−2) was used to study the in vitro evolution of PrPSc from SS21. Unexpectedly, 2 out of the 3 populations propagated at low PrPSc/PrPC were barely detectable after 5 PMCA rounds, i.e., an expected replication factor of ~1014, and eventually became negative in successive rounds (). After 9 PMCA rounds, only 3 populations were positive by WB, after an overall replication factor of 109 for SS21/a and SS21/b and 1024 per SS21/c. Again, an upward shift of PrPres emerged from one population, SS21/a, subjected to high PrPSc/PrPC propagation. The last PMCA round yielding a positive reaction was used to compare the five populations by discriminatory immunoblotting with mAbs SAF84 and SAF32, whose epitope is partially lost in PrPres fragments from vole-adapted scrapie strains.Citation28 This confirmed the upward shift of SS21/a, with SAF84/SAF32 ratio greatly decreased in SS21/a compared with all other SS21-derived populations ( and ).
Figure 3. Evolution of PrPSc in SS21. (A) western blot analysis of PrPres from SS21-derived populations, as indicated. PrPres from different PMCA rounds (roman numbers) were analyzed using antibody SAF84, showing a progressive loss of PrPres in SS21/d and SS21/e and an upward shift in SS21/a. (B) Epitope mapping of PrPres from SS21-derived populations, analyzed in replica blots probed with antibodies SAF84 and SAF32, as indicated. (C) Graph depicting the SAF84/SAF32 antibody ratio (y axis, log scale) for samples shown in panel B. SS21/a had a lower ratio than the other populations, indicating partial retention of the SAF32 epitope.
The same PrPres typing approach was applied to follow PrPSc conformational variations in each of the 9 consecutive PMCA rounds of the mutated SS21/a. This analysis showed that the upward shift of MW and the decrease in SAF84/SAF32 ratio, and so a change in the dominant PrPSc conformational variant, were evident after 2 rounds, but disappeared at the fourth and sixth ones, before being apparently “fixed” by the ninth round (). When this PrPSc variant obtained after 9 PMCA rounds was used as seed in 9 replicate tubes and propagated for 1 additional round, the mutant biochemical signature was lost in 2 out of 9 tubes (), indicating a stochastic preferential propagation of different PrPSc conformers, or a dynamic equilibrium among PrPSc sub-populations with slightly different conformations. In agreement, every attempt to “fix” this conformational variant by end-point dilution failed.
Figure 4. Genesis and evolution of the mutant conformer SS21/a. (A) western blot analysis of PrPres from SS21/a after each PMCA round, analyzed in replica blots probed with antibodies SAF84 and SAF32 as indicated, revealed a fluctuation of the MW in the different rounds of PMCA. The graph showing the SAF84/SAF32 ratio (bottom side of the panel) confirmed that SS21/a IX was different from the inoculum SS21 and identified a fluctuation of the dominant conformational variant among the rounds. (B) SS21/a obtained after 9 PMCA rounds was diluted 1:100, splitted in 9 tubes and subjected to one PMCA round. PrPres was analyzed in replica blots probed with antibodies SAF84 and SAF32, as indicated. The graph in the bottom panel depicts SAF84/SAF32 ratios for samples shown in the blots. PrPres amplified in tubes 8 and 9 showed a loss of the mutant biochemical signature.
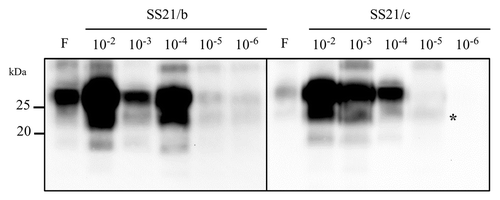
Given the unexpected loss of 2 out of 3 populations propagated at low PrPSc/PrPC, we determined the amplification rates of all the SS21-derived populations by seeding serial 10-fold dilutions of their last positive PMCA round (the ninth for SS21/a, SS21/b and SS21/c and the fourth for SS21/d and SS21/e). In agreement with previous findings (see ), the in vitro amplification of SS21/d and SS21/e was extremely inefficient, as PrPres was barely detectable only from dilution 10−2 (data not shown). Among the 3 populations positive after 9 PMCA rounds, SS21/a and SS21/b showed an amplification rate similar or even higher than that of the starting inoculum. In contrast, SS21/c, the most efficient of the 3 population propagated at low PrPSc/PrPC, was at least 100 times less efficient than those propagated at high PrPSc/PrPC (). Thus, a remarkable loss of fitness was associated with relatively few bottleneck passages in the SS21-derived populations propagated at low PrPSc/PrPC.
Figure 5. Amplification rates. Western blot of PrPres after a single PMCA round of 10-fold dilutions of SS21/b and SS21/c. The band identified by an asterisk in the fifth lane of sample SS21/c represents non fully digested PrPC, often observed in post-PMCA products with low levels of PrPres. PrPres was detected with antibody SAF84.
Discussion
It is now accepted that PMCA is able to reproduce several aspects of prion biology, including prion strain mutation and selection.Citation9,Citation29 It is conceivable that the in vitro environment offers less constraint to prion replication than live animals or cells, due to the absence of active clearance and cell division, which are key players of conformers selection in ex vivo models.Citation17 In agreement, it was shown that PMCA was able to propagate equally well the scrapie strain RML in cell lysates from either, RML-susceptible or RML-resistant cells, thus overcoming cell-specific susceptibility.Citation30
It has been proposed that prion populations are composed of a heterogeneous ensemble of conformational variants which behaves as quasispecies,Citation14 which would imply that mutation is an intrinsic feature of prions. PMCA appeared to us as an appropriate and practical technique to investigate prion mutability in absence of PrP sequence mismatches, PrPC modifications, RNA-depletion or treatments with drugs. Importantly, PMCA allows prion populations to undergo very large amplification factors in a relatively short time period, so that we were able to follow prion populations for a very high number of replications. Indeed, with v586 we observed an increase up to 107 fold of the original population in 48 h, with a much higher kinetic of accumulation than an extremely rapid in vivo model.Citation23 Another key feature of PMCA is that it enables to recover the original population size in a given time period when starting from prion dilutions differing as much as 5 orders of magnitude (see ). We took advantage of this feature to follow prion populations propagated by transferring different population sizes at each PMCA round, mimicking large population vs bottleneck passages, a very useful experimental set up for studying the evolutionary features of quasispecies populations of RNA viruses.Citation18
As a read-out for conformational mutations of PrPSc we used a PrPres typing technique proved to be a reproducible and fast way to reveal even minor changes of PK-cleavage sites of PrPSc.Citation27 On the other hand, PrPres typing only looks for prion variants in which the conformational change involves a shift of the protease-resistant core of PrPSc, which are conceivably much less than the existing PrPSc conformational variants. This necessarily led to underestimate the number of existing variants, notwithstanding we observed the emergence of conformational variants in 1 out of 5 populations with both strains used as starting material. These conformational variants were either positively selected as the dominant population, such as in v586/a, or in dynamic equilibrium with parental variants, as in SS21/a. Interestingly, v586/a and SS21/a mutant populations emerged after a relatively low number of replications, i.e., after 2 large population passages or ~100 fold amplification of the starting population. In contrast, no variations were observed in 6/6 populations subjected to bottleneck events, although these last were grown for a much higher replication factor, up to 1024 for SS21/c. These results might suggest that PrPSc conformational variants with a greater protease-resistant core were present as minor sub-populations in brain-derived v586 and ss21 strains, and were transferred and positively selected by the in vitro environment after large population passages, but instead diluted away in populations propagated at low PrPSc/PrPC ratio. The repeated conformational changes observed in the population SS21/a could thus represent a dynamic equilibrium between two prevalent PrPSc conformations, which could have been positively or negatively selected by uncontrolled variations of the experimental conditions, such as those we have observed while setting up these experiments (Suppl. Figure). An alternative and less realistic explanation would be that prion replication with PMCA is extremely error prone, so to allow mutation and positive selection of the mutant variant after few cycles of replication.
Although no PrPres variants were observed in the prion populations subjected to repeated bottleneck events, throughout our experiments we observed a lower than expected amplification rate in prion populations propagated through serial high dilution passages. Indeed, although v586 yielded as much as PrPSc as in the starting material when seeded at 10−7 dilution, populations serially propagated through 10−7 passages were eventually lost after a few passages. Even when introducing a large population transfer to allow the recovery of population size between 2 bottleneck passages, 2 out of 3 SS21-derived populations became undetectable after 5 PMCA rounds, as their amplification rate had become extremely low after 2 bottleneck passages. SS21/c, whose population size was kept relatively high until the ninth PMCA round, had a ~100 times lower amplification rate than the original population, supporting the view of a generalized fitness decrease in the populations propagated in the bottleneck set up. In sharp contrast, SS21/a and SS21/b, propagated by large population passages, preserved their ability to propagate at high rate.
These findings were surprising in light of recent studies showing an increase in fitness of brain-derived prion strains after 6 PMCA rounds,Citation29 as it would be expected by positive selection of the conformations more fit in the new environment. Why did large population transfers of SS21 allow maintenance of the fitness and accumulation of a conformational variant, while repeated bottleneck events led to a strong decrease of the fitness with no evidence of changes in PrPres type? Interestingly, a possible explanation comes from the quasispecies theory and from studies with RNA viruses supporting the notion that the starting population size affects the evolutionary outcome of a given population under a new selective environment.Citation18 Indeed, populations with high mutation rate are composed by a cloud of variants shaped by environmental constraints, so that sampling a low number of replicative units (bottlenecks) to start a new population implies a loss of “genomic” information compared with large population passages, and thus a loss in adaptability.Citation18 In this scenario, prion populations starting from large number of replicative units might have undergone a rapid accumulation of conformational variants, some of which could have outcompeted the parental conformations by positive selection in the in vitro environment. In contrast, bottleneck events would have decreased the number of conformational variants in the founder population, while increasing the chances of selecting defective variants, with repeated bottleneck events leading to random genetic drift and overall loss of fitness.
In conclusion, our experiments have shown the emergence of mutant PrPSc conformational variants during standard in vitro replication by PMCA. In contrast to previous in vitro studies, in which prion mutants emergences had been forced through PrP sequence mismatchesCitation8 or by changes in the replication environment,Citation9 in our studies prion mutation and selection occurred under constant environmental conditions. Furthermore, we observed a decline of fitness in populations grown through repeated bottlenecks events, which is tentatively explained as random drift of a highly heterogeneous prion population due to repeated founder events. Overall, these findings support the emerging view that mutability is an intrinsic property of prion strains.
Material and Methods
PMCA
Substrates were prepared using 2–3 mo old bank voles homozygous for methionine at codon 109 (Bv109M). Voles were euthanized using carbon dioxide and then perfused with PBS plus 5 mM EDTA. All the brains dissected were immediately homogenized in Conversion Buffer (PBS 1×, pH 7,4; 0,15 M NaCl; 1% Triton X) with the Roche Complete Protease Inibitor Cocktail (1 tablet in 50 ml CB) as 10% w/v; the pool obtained was divided into small aliquots and stored at -80 °C. All the procedures for substrate preparation were conducted in a prion free laboratory, and the aliquots needed were taken in the prion lab only immediately before the beginning of each PMCA round.
Seeds were prepared using brain tissue from Bv109M terminally affected with strains SS21 or v586. Tissues were homogenized in PBS (10% w/v) containing Complete Protease Inibitor Cocktail (Roche) using disposable Teflon pestles directly in 1.5 mL Eppendorf tubes, and stored at -20 °C. This homogenate was serially diluted 10-fold in normal bank vole homogenate to generate a dilution curve from 10−2 to 10−8 (where 10−2 means 1% w/v of infected brain homogenate) using the procedure described in Cosseddu et al.,Citation24 to avoid contamination.
PMCA was performed using the Misonix S3000 sonicator, with procedures specifically designed to avoid cross-contamination of samples as described in Cosseddu et al.Citation24 Briefly, to avoid cross-contamination within the PMCA machine 0.5 mL screw cap Multiply-Safecup (Sarstedt) sealed with parafilm were used, and we avoided to work simultaneously with the 2 strains either in the sonicator rack or under the biological hood. At the end of each round, 5 μl of each reaction mix were serially 10-fold diluted in fresh substrate and the needed dilution subjected to the new PMCA round, according to the experimental design depicted in . The amplification factor was determined as the reciprocal of the dilution factor that a given population accumulated during PMCA passages.
Western blot analysis
Fifteen μl of seeds or PMCA reactions were added to 15 μl of TRIS-HCl sarcosyl 4% and digested with 100 μg/mL of proteinase K (Sigma-Aldrich) for diagnosis or with 200 μg/ml of proteinase K for epitope mapping. Electrophoresis and western blotting were performed as previously described.Citation28 The membrane were then analyzed with anti-PrP monoclonal antibody SAF84 (a.a. 167–173; 1.2 μg/ml), 12B2 (a.a. 93–97; 0.4 μg/ml), and SAF32 (octarepeat; 4.8 μg/ml). Following incubation with horseradish peroxidase-conjugated anti-mouse immunoglobulin (Pierce Biotechnology) at 1:13 000, the PrP bands were detected by enhanced chemiluminescent substrate (SuperSignal Femto, Pierce) and VersaDoc imaging system (Bio-Rad). The chemiluminescence signal was quantified by QuantityOne software (Bio-Rad).
Additional material
Download Zip (182.5 KB)Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was supported by the Italian Ministry of Health (RF-2009–1474624).
Supplemental Materials
Supplemental materials may be found here:
https://www.landesbioscience.com/journals/prion/article/28468/
References
- Saborio GP, Permanne B, Soto C. Sensitive detection of pathological prion protein by cyclic amplification of protein misfolding. Nature 2001; 411:810 - 3; http://dx.doi.org/10.1038/35081095; PMID: 11459061
- Castilla J, Saá P, Hetz C, Soto C. In vitro generation of infectious scrapie prions. Cell 2005; 121:195 - 206; http://dx.doi.org/10.1016/j.cell.2005.02.011; PMID: 15851027
- Bessen RA, Marsh RF. Biochemical and physical properties of the prion protein from two strains of the transmissible mink encephalopathy agent. J Virol 1992; 66:2096 - 101; PMID: 1347795
- Telling GC, Parchi P, DeArmond SJ, Cortelli P, Montagna P, Gabizon R, Mastrianni J, Lugaresi E, Gambetti P, Prusiner SB. Evidence for the conformation of the pathologic isoform of the prion protein enciphering and propagating prion diversity. Science 1996; 274:2079 - 82; http://dx.doi.org/10.1126/science.274.5295.2079; PMID: 8953038
- Safar J, Wille H, Itri V, Groth D, Serban H, Torchia M, Cohen FE, Prusiner SB. Eight prion strains have PrP(Sc) molecules with different conformations. Nat Med 1998; 4:1157 - 65; http://dx.doi.org/10.1038/2654; PMID: 9771749
- Collinge J, Clarke AR. A general model of prion strains and their pathogenicity. Science 2007; 318:930 - 6; http://dx.doi.org/10.1126/science.1138718; PMID: 17991853
- Bruce ME. TSE strain variation. Br Med Bull 2003; 66:99 - 108; http://dx.doi.org/10.1093/bmb/66.1.99; PMID: 14522852
- Castilla J, Gonzalez-Romero D, Saá P, Morales R, De Castro J, Soto C. Crossing the species barrier by PrP(Sc) replication in vitro generates unique infectious prions. Cell 2008; 134:757 - 68; http://dx.doi.org/10.1016/j.cell.2008.07.030; PMID: 18775309
- Gonzalez-Montalban N, Lee YJ, Makarava N, Savtchenko R, Baskakov IV. Changes in prion replication environment cause prion strain mutation. FASEB J 2013; 27:3702 - 10; http://dx.doi.org/10.1096/fj.13-230466; PMID: 23729586
- Makarava N, Savtchenko R, Baskakov IV. Selective amplification of classical and atypical prions using modified protein misfolding cyclic amplification. J Biol Chem 2013; 288:33 - 41; http://dx.doi.org/10.1074/jbc.M112.419531; PMID: 23168413
- Angers RC, Kang HE, Napier D, Browning S, Seward T, Mathiason C, Balachandran A, McKenzie D, Castilla J, Soto C, et al. Prion strain mutation determined by prion protein conformational compatibility and primary structure. Science 2010; 328:1154 - 8; http://dx.doi.org/10.1126/science.1187107; PMID: 20466881
- Mahal SP, Baker CA, Demczyk CA, Smith EW, Julius C, Weissmann C. Prion strain discrimination in cell culture: the cell panel assay. Proc Natl Acad Sci U S A 2007; 104:20908 - 13; http://dx.doi.org/10.1073/pnas.0710054104; PMID: 18077360
- Weissmann C. Mutation and selection of prions. PLoS Pathog 2012; 8:e1002582; http://dx.doi.org/10.1371/journal.ppat.1002582; PMID: 22479179
- Li J, Browning S, Mahal SP, Oelschlegel AM, Weissmann C. Darwinian evolution of prions in cell culture. Science 2010; 327:869 - 72; http://dx.doi.org/10.1126/science.1183218; PMID: 20044542
- Mahal SP, Browning S, Li J, Suponitsky-Kroyter I, Weissmann C. Transfer of a prion strain to different hosts leads to emergence of strain variants. Proc Natl Acad Sci U S A 2010; 107:22653 - 8; http://dx.doi.org/10.1073/pnas.1013014108; PMID: 21156827
- Weissmann C, Li J, Mahal SP, Browning S. Prions on the move. EMBO Rep 2011; 12:1109 - 17; http://dx.doi.org/10.1038/embor.2011.192; PMID: 21997298
- Oelschlegel AM, Weissmann C. Acquisition of drug resistance and dependence by prions. PLoS Pathog 2013; 9:e1003158; http://dx.doi.org/10.1371/journal.ppat.1003158; PMID: 23408888
- Domingo E, Sheldon J, Perales C. Viral quasispecies evolution. Microbiol Mol Biol Rev 2012; 76:159 - 216; http://dx.doi.org/10.1128/MMBR.05023-11; PMID: 22688811
- Bateman DA, Wickner RB. The [PSI+] prion exists as a dynamic cloud of variants. PLoS Genet 2013; 9:e1003257; http://dx.doi.org/10.1371/journal.pgen.1003257; PMID: 23382698
- Nonno R, Di Bari MA, Cardone F, Vaccari G, Fazzi P, Dell’Omo G, Cartoni C, Ingrosso L, Boyle A, Galeno R, et al. Efficient transmission and characterization of Creutzfeldt-Jakob disease strains in bank voles. PLoS Pathog 2006; 2:e12; http://dx.doi.org/10.1371/journal.ppat.0020012; PMID: 16518470
- Cartoni C, Schininà ME, Maras B, Nonno R, Vaccari G, Di Bari M, Conte M, De Pascalis A, Principe S, Cardone F, et al. Quantitative profiling of the pathological prion protein allotypes in bank voles by liquid chromatography-mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci 2007; 849:302 - 6; http://dx.doi.org/10.1016/j.jchromb.2006.08.016; PMID: 17008136
- Di Bari MA, Chianini F, Vaccari G, Esposito E, Conte M, Eaton SL, Hamilton S, Finlayson J, Steele PJ, Dagleish MP, et al. The bank vole (Myodes glareolus) as a sensitive bioassay for sheep scrapie. J Gen Virol 2008; 89:2975 - 85; http://dx.doi.org/10.1099/vir.0.2008/005520-0; PMID: 19008382
- Di Bari MA, Nonno R, Castilla J, D’Agostino C, Pirisinu L, Riccardi G, Conte M, Richt J, Kunkle R, Langeveld J, et al. Chronic wasting disease in bank voles: characterisation of the shortest incubation time model for prion diseases. PLoS Pathog 2013; 9:e1003219; http://dx.doi.org/10.1371/journal.ppat.1003219; PMID: 23505374
- Cosseddu GM, Nonno R, Vaccari G, Bucalossi C, Fernandez-Borges N, Di Bari MA, Castilla J, Agrimi U. Ultra-efficient PrP(Sc) amplification highlights potentialities and pitfalls of PMCA technology. PLoS Pathog 2011; 7:e1002370; http://dx.doi.org/10.1371/journal.ppat.1002370; PMID: 22114554
- Pirisinu L, Di Bari M, Marcon S, Vaccari G, D’Agostino C, Fazzi P, Esposito E, Galeno R, Langeveld J, Agrimi U, et al. A new method for the characterization of strain-specific conformational stability of protease-sensitive and protease-resistant PrP. PLoS One 2010; 5:e12723; http://dx.doi.org/10.1371/journal.pone.0012723; PMID: 20856860
- Pirisinu L, Migliore S, Di Bari MA, Esposito E, Baron T, D’Agostino C, De Grossi L, Vaccari G, Agrimi U, Nonno R. Molecular discrimination of sheep bovine spongiform encephalopathy from scrapie. Emerg Infect Dis 2011; 17:695 - 8; http://dx.doi.org/10.3201/eid1704.101215; PMID: 21470463
- Migliore S, Esposito E, Pirisinu L, Marcon S, Di Bari M, D’Agostino C, Chiappini B, Conte M, Sezzi E, De Grossi L, et al. Effect of PrP genotype and route of inoculation on the ability of discriminatory Western blot to distinguish scrapie from sheep bovine spongiform encephalopathy. J Gen Virol 2012; 93:450 - 5; http://dx.doi.org/10.1099/vir.0.035469-0; PMID: 21994325
- Pirisinu L, Marcon S, Di Bari M, D'Agostino C, Agrimi U, Nonno R. Biochemical characterization of prion strains in bank voles. Pathogens 2013; 2:446 - 56; http://dx.doi.org/10.3390/pathogens2030446
- Gonzalez-Montalban N, Baskakov IV. Assessment of strain-specific PrP(Sc) elongation rates revealed a transformation of PrP(Sc) properties during protein misfolding cyclic amplification. PLoS One 2012; 7:e41210; http://dx.doi.org/10.1371/journal.pone.0041210; PMID: 22815972
- Herva ME, Weissman C. Cell-specific susceptibility to prion strains is a property of the intact cell. Prion 2012; 6:371 - 4; http://dx.doi.org/10.4161/pri.20198; PMID: 22561192
