Abstract
Self-assembly of proteins and peptides into amyloid structures has been the subject of intense and focused research due to their association with neurodegenerative, age-related human diseases and transmissible prion diseases in humans and mammals. Of the disease associated amyloid assemblies, a diverse array of species, ranging from small oligomeric assembly intermediates to fibrillar structures, have been shown to have toxic potential. Equally, a range of species formed by the same disease associated amyloid sequences have been found to be relatively benign under comparable monomer equivalent concentrations and conditions. In recent years, an increasing number of functional amyloid systems have also been found. These developments show that not all amyloid structures are generically toxic to cells. Given these observations, it is important to understand why amyloid structures may encode such varied toxic potential despite sharing a common core molecular architecture. Here, we discuss possible links between different aspects of amyloidogenic structures and assembly mechanisms with their varied functional effects. We propose testable hypotheses for the relationship between amyloid structure and its toxic potential in the context of recent reports on amyloid sequence, structure, and toxicity relationships.
From Sequence to Amyloid Structure
Amyloid fibrils are highly organized and stable structures composed of proteins that are able to self-assemble via β-sheet association. The core of all amyloid fibrils adopt what is commonly known as the cross-β structure, named due to the arrangement in which β-strands that run perpendicular to the fiber axis form β-sheet ribbons or filaments that further associate to form amyloid fibrils.Citation1 These fibrils are found in a large number of protein misfolding diseases including Alzheimer disease, type 2 diabetes mellitus, and transmissible spongiform encephalopathies.Citation2 Peptides and proteins with different sequences are able to readily form amyloid-like fibrils. Thus, the sequence determinants of amyloid self-assembly have been an area of intense research. Algorithms have been developed to analyze amino acid sequences and to predict their propensity to form amyloid. These have used properties such as β-sheet propensity and hydrophobicityCitation3,Citation4 or more complex methods to look at the relationship between the arrangement of the amino acids and the final cross-β structure.Citation5,Citation6 From these studies, it has become clear that many short peptides have a high propensity to form amyloid fibrilsCitation6; over 50 have been identified to date and the number is growing. Recent advances in characterizing the structural properties of amyloid fibrils show an increasingly detailed atomic resolution picture of the cross-β structure. X-ray crystallography provides structural information, showing that small amyloid forming peptides are able to form steric zippers in which side chains associate and interlock between the β-sheets.Citation7 X-ray fiber diffractionCitation8,Citation9 and solid-state NMRCitation10-Citation12 have been used to examine the structure of the fibrils themselves, and show similar complementarity between the side chain interactions across the sheets. These observations further anchor the notion of a similar core structural organization for all amyloid fibrils ().
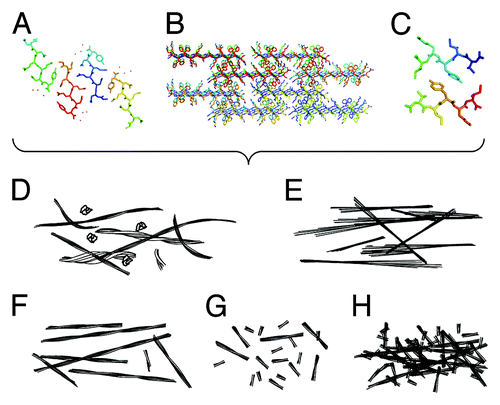
Figure 1. Detailed atomic resolution structures of amyloid leads to different physical properties, which in turn produce varied biological functions. (A–C) Structural models depicting the amyloid cross-β molecular architecture. (A) the crystal structure of GNNQQNY (2OMM.pdb),Citation7 (B) a model of fibrous nanocrystals formed by KFFEAAAKKFFE,Citation8 (C) a model structure of fibrils formed by VIYKI.Citation9 (D–H) schematic illustrations of morphologies that may result in different biological properties. (D) Illustrations of flexible and heterogeneous fibrils compared with (E) rigid and crystal-like fibrils. (F) Illustrations showing long and thermally, mechanically and/or enzymatically stable fibrils compared with (G) short, fragile, and unstable fibrils with non-interacting surfaces and (H) fibrils with interacting or sticky surfaces.
Toxic and Functional Properties of Amyloid
In amyloid associated disease, amyloid fibrils are deposited in tissues, forming insoluble structures and sometimes replacing cells. For example, in lysozyme amyloidosis, much of the kidney can be replaced by amyloid deposits.Citation13 However, some small oligomeric intermediates formed during the self-assembly by amyloidogenic precursor proteins or peptides have toxic potential and are involved in the cell death observed in these misfolding and neurodegenerative diseases,Citation14 although not all small oligomeric intermediates have been found to be toxic. For example, HypF can be induced to form both toxic and non-toxic oligomeric species.Citation15 Equally, small fibrillar oligomer species or fibril fragments have also been found to possess toxic potential.Citation16-Citation18 Indeed, a β-sheet protein that has been designed to form amyloid-like oligomers leads to a cascade of toxic events following expression in human cells.Citation17 Recently, it has become clear that amyloid structures are quite commonly found in natural systems, and there are increasing reports of such functional amyloid structures that provide scaffold, support, and protection due to their stability and strength.Citation19 For example, E. coli produces curli that forms a layer of amyloid fibrils on the outside of the organism,Citation20 while amyloid has also been identified as forming a protective layer to melanosomes.Citation21 Interestingly, the self-assembly of each of these systems is carefully controlled by the organism,Citation22 although whether such functional amyloid assemblies inherently lack toxic potential remains to be established.
These observations together with recent advances in the determination of the sequence and structural properties of amyloid fibrils raise an important question: Why are some amyloid assemblies toxic, while others are inert or tolerated, even to the extent that they may be evolved to perform useful biological functions, despite the fact that the end products of amyloid assembly share a common cross-β structural organization?
In Search of Structure and Assembly Properties that Confer Toxic Potential
The varied toxic potential of amyloid assemblies may involve several structural and assembly properties such as different levels of structural rigidity, stability (thermal, enzymatic, or mechanical), and surface protection of amyloid fibrils (). These form testable hypotheses that can be examined by studies that connect sequence, structure, morphology, and toxicity of amyloid assemblies.
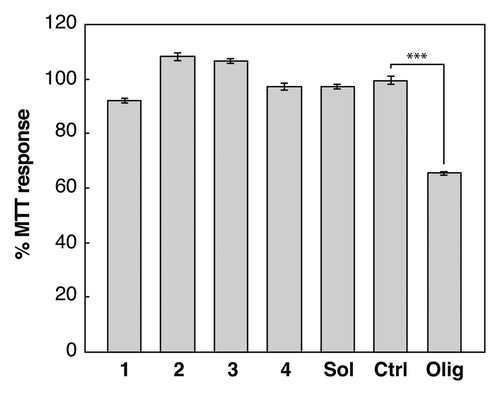
Some short amyloidogenic peptides identified by algorithms and crystallography, are some of the most efficient and effective amyloid fibril formers. They self-assemble rapidly and effectively, forming highly organized and rigid fibrils and sometimes crystal-like aggregatesCitation23 (). Using these highly effective self-assembling peptides, we investigated the cytotoxic properties of some of these rigid crystal-like assemblies in neuroblastoma cell-line cultures, compared with toxic, oligomeric Aβ(1−42) (Aβ42) (). We investigated the effect of soluble peptide as well as crystalline and fibrillar aggregates formed by the peptide KFFEAAAKKFFE and variants of this sequence.Citation24 Interestingly, none affected the reduction of MTT to formazan, a process that occurs in viable cells with active metabolism, suggesting that these aggregates possess little cytotoxic potential. For comparison, it is well known that Aβ, in various forms, is toxic to cells. Aβ peptides self-assemble rapidly,Citation25 particularly Aβ42. However, Aβ42 does form significant populations of potentially long-lived and toxic intermediates that can be visualized by electron microscopy.Citation16,Citation26 Aβ also has the potential to form a wide range of different species and fibril polymorphs.Citation27,Citation28 Thus, the lack of structural rigidity in the fibrillar assembly products of Aβ amyloid may reflect the ability of Aβ to participate in heterogeneous assembly reactions that could form a large number of different intermediate and fibril species, some of which may possess cytotoxic potential.
Figure 2. Structure and morphology of different amyloid aggregates assembled from KFFEAAAKKFFE Citation8 and variants Citation24 results in altered morphology and assembly properties. Images show aggregated peptide samples 1: KFFEAAAKKFFE in PBS, 2: RFFEAAAKRFFE in PBS, 3: AFFEAAAAKFFE in water, and 4: KFFEAAAAKFFE in water. Upper row shows negative stain transmission electron microscope images (black scale bars = 200 nm). Lower row shows atomic force microscopy height images (white scale bars = 2 μm).
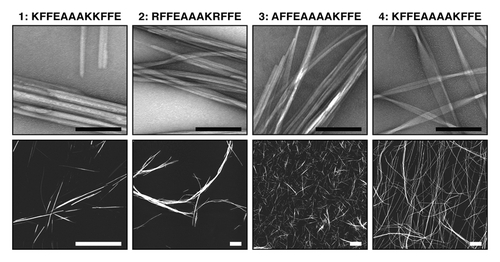
Figure 3. The effect of assembled peptide morphology and composition on neuroblastoma cells. MTT assays (Molecular Probes) were performed according to the manufacturers protocol. A neuroblastoma cell line (SH-SY5Y) was treated with 10 μM monomer equivalent concentrations of aggregated peptide samples 1: KFFEAAAKKFFE in PBS 2: RFFEAAAKRFFE in PBS, 3: AFFEAAAAKFFE in water, and 4: KFFEAAAAKFFE in water (). Cells were also treated with soluble KFFEAAAKKFFE in water (labeled Sol), a buffer only control (labeled Ctrl) or Aβ42 oligomers (labeled Olig). Changes in the MTT response after 24 h were monitored and are shown relative to the buffer control and error bars correspond to SEM over 3 replicates. Only Aβ42 oligomersCitation16,Citation26 induce cellular dysfunction resulting in a reduction to 60% compared with control. The student t test comparison between olig and ctrl showed significance ofP < 0.001 labeled ***.
Another property to consider is the stability of amyloid fibrils, and their reversibility and disassembly processes. The assembly process of amyloid is reversible, and this dynamic process is normally favored toward the formation of amyloid fibrils. Thus, fibrils that are thermodynamically and/or kinetically more stable will disassemble more slowly and, therefore, shed a lower population of potentially cytotoxic speciesCitation29 from fibril ends at a lower rate. The mechanical stability of amyloid fibrils may also play a key role in generating small fibril fragments (or fibrillar oligomers) that possess cytotoxic potential through fibril breakage processes induced by mechanical or enzymatic action.Citation30,Citation31 Thus, understanding the origin of the toxic properties of amyloid assemblies will involve the characterization of the stability of fibrils toward different perturbations, and the sequence and structural properties that confer the different stabilities of fibrils.
The surface properties of amyloid fibrils may also influence their toxic potential. For example, small oligomeric species and fibrillar fragments have all been shown to be able to disrupt lipid membranes.Citation18,Citation32,Citation33 For fibril fragments, fibril ends have been seen as the active surfaces that interact with membranes.Citation32-Citation34 Thus, the lack of surface protection that can minimize aberrant interactions at the sites of amyloid growth may play an important role in terms of their cytotoxic potential. Such protection, for example in the form of a mechanism involving capping of growth competent fibril ends, may also be a generic feature that provides a host organism the means to control functional amyloid. Surfaces that promote fibril-fibril interactions, or the packing between proto-filaments in fibrils may further modulate the reactivity and stability of amyloid fibrils, deposits, or plaques, in turn affecting the generation of potentially toxic species through depolymerisation. Detailed atomic resolution structural information of amyloid fibrils may inform on their surface properties, for example revealing hydrophobic patches that may act as “sticky” sites that can promote aberrant interactions or fibril clumping.
Amyloid by Design
Finally, a large area of synthetic biology has grown from the use of β-sheet structural growth, providing nanowires and hydrogels composed of amyloid-like structures.Citation11,Citation35,Citation36 The amyloid structure may prove to be useful and may be exploited to form a range of materials.Citation37 These fibrils are formed by repetitive precursor sequences, often short peptides, that form highly organized fibrils. These can be decorated further, or they can be used to form hydrogels or wires.Citation38,Citation39 β-sheet based materials have been used in tissue engineering and for enabling cell growth, showing they are non-toxic.Citation35
Ultimately, engineering synthetic amyloid as a novel and safe bionanomaterial will rely on the understanding of the structural factors that underpin toxic properties so that the toxic potential of designed amyloid materials can be minimized or avoided. In the same way, understanding the same underlying principles will be key in understanding amyloid disease so that the toxic potential of disease associated amyloid can be inhibited.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
L.C.S. and K.E.M. acknowledge funding from MRC MR/K022105/1 and Alzheimer research UK. R.M. and W.F.X. acknowledge funding from BBSRC BB/J008001/1.
References
- Morris KL, Serpell LC. X-ray fibre diffraction studies of amyloid fibrils. Methods Mol Biol 2012; 849:121 - 35; http://dx.doi.org/10.1007/978-1-61779-551-0_9; PMID: 22528087
- Bucciantini M, Giannoni E, Chiti F, Baroni F, Formigli L, Zurdo J, Taddei N, Ramponi G, Dobson CM, Stefani M. Inherent toxicity of aggregates implies a common mechanism for protein misfolding diseases. Nature 2002; 416:507 - 11; http://dx.doi.org/10.1038/416507a; PMID: 11932737
- Pawar AP, Dubay KF, Zurdo J, Chiti F, Vendruscolo M, Dobson CM. Prediction of “aggregation-prone” and “aggregation-susceptible” regions in proteins associated with neurodegenerative diseases. J Mol Biol 2005; 350:379 - 92; http://dx.doi.org/10.1016/j.jmb.2005.04.016; PMID: 15925383
- Zibaee S, Jakes R, Fraser G, Serpell LC, Crowther RA, Goedert M. Sequence Determinants for Amyloid Fibrillogenesis of Human alpha-Synuclein. J Mol Biol 2007; 374:454 - 64; http://dx.doi.org/10.1016/j.jmb.2007.09.039; PMID: 17936783
- Thompson MJ, Sievers SA, Karanicolas J, Ivanova MI, Baker D, Eisenberg D. The 3D profile method for identifying fibril-forming segments of proteins. Proc Natl Acad Sci U S A 2006; 103:4074 - 8; http://dx.doi.org/10.1073/pnas.0511295103; PMID: 16537487
- Maurer-Stroh S, Debulpaep M, Kuemmerer N, Lopez de la Paz M, Martins IC, Reumers J, Morris KL, Copland A, Serpell L, Serrano L, et al. Exploring the sequence determinants of amyloid structure using position-specific scoring matrices. Nat Methods 2010; 7:237 - 42; http://dx.doi.org/10.1038/nmeth.1432; PMID: 20154676
- Sawaya MR, Sambashivan S, Nelson R, Ivanova MI, Sievers SA, Apostol MI, Thompson MJ, Balbirnie M, Wiltzius JJ, McFarlane HT, et al. Atomic structures of amyloid cross-beta spines reveal varied steric zippers. Nature 2007; 447:453 - 7; http://dx.doi.org/10.1038/nature05695; PMID: 17468747
- Makin OS, Atkins E, Sikorski P, Johansson J, Serpell LC. Molecular basis for amyloid fibril formation and stability. Proc Natl Acad Sci U S A 2005; 102:315 - 20; http://dx.doi.org/10.1073/pnas.0406847102; PMID: 15630094
- Morris KL, Rodger A, Hicks MR, Debulpaep M, Schymkowitz J, Rousseau F, Serpell LC. Exploring the sequence-structure relationship for amyloid peptides. Biochem J 2013; 450:275 - 83; http://dx.doi.org/10.1042/BJ20121773; PMID: 23252554
- Lu JX, Qiang W, Yau WM, Schwieters CD, Meredith SC, Tycko R. Molecular structure of β-amyloid fibrils in Alzheimer’s disease brain tissue. Cell 2013; 154:1257 - 68; http://dx.doi.org/10.1016/j.cell.2013.08.035; PMID: 24034249
- Fitzpatrick AW, Debelouchina GT, Bayro MJ, Clare DK, Caporini MA, Bajaj VS, Jaroniec CP, Wang L, Ladizhansky V, Müller SA, et al. Atomic structure and hierarchical assembly of a cross-β amyloid fibril. Proc Natl Acad Sci U S A 2013; 110:5468 - 73; http://dx.doi.org/10.1073/pnas.1219476110; PMID: 23513222
- Van Melckebeke H, Wasmer C, Lange A, Ab E, Loquet A, Böckmann A, Meier BH. Atomic-resolution three-dimensional structure of HET-s(218-289) amyloid fibrils by solid-state NMR spectroscopy. J Am Chem Soc 2010; 132:13765 - 75; http://dx.doi.org/10.1021/ja104213j; PMID: 20828131
- Pepys MB, Hawkins PN, Booth DR, Vigushin DM, Tennent GA, Soutar AK, Totty N, Nguyen O, Blake CC, Terry CJ, et al. Human lysozyme gene mutations cause hereditary systemic amyloidosis. Nature 1993; 362:553 - 7; http://dx.doi.org/10.1038/362553a0; PMID: 8464497
- Irvine GB, El-Agnaf OM, Shankar GM, Walsh DM. Protein aggregation in the brain: the molecular basis for Alzheimer’s and Parkinson’s diseases. Mol Med 2008; 14:451 - 64; http://dx.doi.org/10.2119/2007-00100.Irvine; PMID: 18368143
- Relini A, Torrassa S, Ferrando R, Rolandi R, Campioni S, Chiti F, Gliozzi A. Detection of populations of amyloid-like protofibrils with different physical properties. Biophys J 2010; 98:1277 - 84; http://dx.doi.org/10.1016/j.bpj.2009.11.052; PMID: 20371327
- Williams TL, Day IJ, Serpell LC. The effect of Alzheimer’s Aβ aggregation state on the permeation of biomimetic lipid vesicles. Langmuir 2010; 26:17260 - 8; http://dx.doi.org/10.1021/la101581g; PMID: 20923185
- Olzscha H, Schermann SM, Woerner AC, Pinkert S, Hecht MH, Tartaglia GG, Vendruscolo M, Hayer-Hartl M, Hartl FU, Vabulas RM. Amyloid-like aggregates sequester numerous metastable proteins with essential cellular functions. Cell 2011; 144:67 - 78; http://dx.doi.org/10.1016/j.cell.2010.11.050; PMID: 21215370
- Xue WF, Hellewell AL, Gosal WS, Homans SW, Hewitt EW, Radford SE. Fibril fragmentation enhances amyloid cytotoxicity. J Biol Chem 2009; 284:34272 - 82; http://dx.doi.org/10.1074/jbc.M109.049809; PMID: 19808677
- Fowler DM, Koulov AV, Balch WE, Kelly JW. Functional amyloid--from bacteria to humans. Trends Biochem Sci 2007; 32:217 - 24; http://dx.doi.org/10.1016/j.tibs.2007.03.003; PMID: 17412596
- Wang X, Hammer ND, Chapman MR. The molecular basis of functional bacterial amyloid polymerization and nucleation. J Biol Chem 2008; 283:21530 - 9; http://dx.doi.org/10.1074/jbc.M800466200; PMID: 18508760
- Fowler DM, Koulov AV, Alory-Jost C, Marks MS, Balch WE, Kelly JW. Functional amyloid formation within mammalian tissue. PLoS Biol 2006; 4:e6; http://dx.doi.org/10.1371/journal.pbio.0040006; PMID: 16300414
- Hammer ND, Schmidt JC, Chapman MR. The curli nucleator protein, CsgB, contains an amyloidogenic domain that directs CsgA polymerization. Proc Natl Acad Sci U S A 2007; 104:12494 - 9; http://dx.doi.org/10.1073/pnas.0703310104; PMID: 17636121
- Marshall K, Serpell LC. Fibres, crystals and polymorphism: the structural promiscuity of amyloidogenic peptides. Soft Matter 2010; 6:2110 - 3; http://dx.doi.org/10.1039/b926623b
- Marshall KE, Morris KL, Charlton D, O’Reilly N, Lewis L, Walden H, Serpell LC. Hydrophobic, aromatic, and electrostatic interactions play a central role in amyloid fibril formation and stability. Biochemistry 2011; 50:2061 - 71; http://dx.doi.org/10.1021/bi101936c; PMID: 21288003
- Hellstrand E, Boland B, Walsh DM, Linse S. Amyloid β-protein aggregation produces highly reproducible kinetic data and occurs by a two-phase process. ACS Chem Neurosci 2010; 1:13 - 8; http://dx.doi.org/10.1021/cn900015v; PMID: 22778803
- Soura V, Stewart-Parker M, Williams TL, Ratnayaka A, Atherton J, Gorringe K, Tuffin J, Darwent E, Rambaran R, Klein W, et al. Visualization of co-localization in Aβ42-administered neuroblastoma cells reveals lysosome damage and autophagosome accumulation related to cell death. Biochem J 2012; 441:579 - 90; http://dx.doi.org/10.1042/BJ20110749; PMID: 21955321
- Sachse C, Grigorieff N, Fändrich M. Nanoscale flexibility parameters of Alzheimer amyloid fibrils determined by electron cryo-microscopy. Angew Chem Int Ed Engl 2010; 49:1321 - 3; http://dx.doi.org/10.1002/anie.200904781; PMID: 20069616
- Paravastu AK, Leapman RD, Yau WM, Tycko R. Molecular structural basis for polymorphism in Alzheimer’s beta-amyloid fibrils. Proc Natl Acad Sci U S A 2008; 105:18349 - 54; http://dx.doi.org/10.1073/pnas.0806270105; PMID: 19015532
- Cremades N, Cohen SI, Deas E, Abramov AY, Chen AY, Orte A, Sandal M, Clarke RW, Dunne P, Aprile FA, et al. Direct observation of the interconversion of normal and toxic forms of α-synuclein. Cell 2012; 149:1048 - 59; http://dx.doi.org/10.1016/j.cell.2012.03.037; PMID: 22632969
- Xue WF, Hellewell AL, Hewitt EW, Radford SE. Fibril fragmentation in amyloid assembly and cytotoxicity: when size matters. Prion 2010; 4:20 - 5; http://dx.doi.org/10.4161/pri.4.1.11378; PMID: 20305394
- Xue WF, Radford SE. An imaging and systems modeling approach to fibril breakage enables prediction of amyloid behavior. Biophys J 2013; 105:2811 - 9; http://dx.doi.org/10.1016/j.bpj.2013.10.034; PMID: 24359753
- Butterfield SM, Lashuel HA. Amyloidogenic protein-membrane interactions: mechanistic insight from model systems. Angew Chem Int Ed Engl 2010; 49:5628 - 54; http://dx.doi.org/10.1002/anie.200906670; PMID: 20623810
- Williams TL, Serpell LC. Membrane and surface interactions of Alzheimer’s Aβ peptide--insights into the mechanism of cytotoxicity. FEBS J 2011; 278:3905 - 17; http://dx.doi.org/10.1111/j.1742-4658.2011.08228.x; PMID: 21722314
- Milanesi L, Sheynis T, Xue WF, Orlova EV, Hellewell AL, Jelinek R, Hewitt EW, Radford SE, Saibil HR. Direct three-dimensional visualization of membrane disruption by amyloid fibrils. Proc Natl Acad Sci U S A 2012; 109:20455 - 60; http://dx.doi.org/10.1073/pnas.1206325109; PMID: 23184970
- Kretsinger JK, Haines LA, Ozbas B, Pochan DJ, Schneider JP. Cytocompatibility of self-assembled beta-hairpin peptide hydrogel surfaces. Biomaterials 2005; 26:5177 - 86; http://dx.doi.org/10.1016/j.biomaterials.2005.01.029; PMID: 15792545
- Yan H, Nykanen A, Ruokolainen J, Farrar D, Gough JE, Saiani A, Miller AF. Thermo-reversible protein fibrillar hydrogels as cell scaffolds. Faraday Discuss 2008; 139:71 - 84, discussion 105-28, 419-20; http://dx.doi.org/10.1039/b717748h; PMID: 19048991
- Rajagopal K, Schneider JP. Self-assembling peptides and proteins for nanotechnological applications. Curr Opin Struct Biol 2004; 14:480 - 6; http://dx.doi.org/10.1016/j.sbi.2004.06.006; PMID: 15313243
- Schneider JP, Pochan DJ, Ozbas B, Rajagopal K, Pakstis L, Kretsinger J. Responsive hydrogels from the intramolecular folding and self-assembly of a designed peptide. J Am Chem Soc 2002; 124:15030 - 7; http://dx.doi.org/10.1021/ja027993g; PMID: 12475347
- Yan H, Saiani A, Gough JE, Miller AF. Thermoreversible protein hydrogel as cell scaffold. Biomacromolecules 2006; 7:2776 - 82; http://dx.doi.org/10.1021/bm0605560; PMID: 17025352
