Abstract
The disease-associated prion protein (PrP) forms aggregates which vary in structural conformation yet share an identical primary sequence. These variations in PrP conformation are believed to manifest in prion strains exhibiting distinctly different periods of disease incubation as well as regionally specific aggregate deposition within the brain. The anionic luminescent conjugated polythiophene (LCP), polythiophene acetic acid (PTAA) has previously been used to distinguish PrP deposits associated with distinct mouse adapted strains via distinct fluorescence emission profiles from the dye. Here, we employed PTAA and 3 structurally related chemically defined luminescent conjugated oligothiophenes (LCOs) to stain brain tissue sections from mice inoculated with 2 distinct prion strains. Our results showed that in addition to emission spectra, excitation, and fluorescence lifetime imaging microscopy (FLIM) can fruitfully be assessed for optical distinction of PrP deposits associated with distinct prion strains. Our findings support the theory that alterations in LCP/LCO fluorescence are due to distinct conformational restriction of the thiophene backbone upon interaction with PrP aggregates associated with distinct prion strains. We foresee that LCP and LCO staining in combination with multimodal fluorescence microscopy might aid in detecting structural differences among discrete protein aggregates and in linking protein conformational features with disease phenotypes for a variety of neurodegenerative proteinopathies.
Abbreviations
| AD | = | Alzheimer disease |
| LCOs | = | luminescent conjugated oligothiophenes |
| LCPs | = | luminescent conjugated polythiophenes |
| mSS | = | murine sheep scrapie |
| mCWD | = | murine chronic wasting disease |
| PrP | = | prion protein |
Introduction
Prion diseases are infectious and fatal neurodegenerative disorders. A common pathological hallmark for these diseases is accumulation of insoluble protein aggregates composed of PrP within the brains of both humans and animals.Citation1-3 Congenic animals infected by prions from different disease origin results in various clinical phenotypes. Conspicuously, conformational variations in the tertiary and/or quaternary structure of PrP aggregates are assumed to give rise to distinct prion strains with different periods of disease incubation, lesion profiles, as well as a regionally specific protein aggregate deposition within the brain.Citation4-7 Recently, it has also been shown that protein aggregates associated with a variety of neurodegenerative diseases, such as a Alzheimer disease (AD) as well as synucleinopathies and tauopathies, exist in multiple aggregated conformations.Citation8-12 Hence, a phenomenon similar to prion strains is emerging in many if not all neurodegenerative proteinopathies, and techniques for distinguishing different aggregate conformations are thus of great importance.
Prions strains were originally distinguished by characteristic incubation periods in differentially susceptible inbred mice,Citation4 but a variety of biochemical techniques can also be used to assess different strains since PrPSc from distinct prion strains can differ in electrophoretic mobility,Citation13 immunological reactivity to antibodies upon proteinase K digestion,Citation14 and relative glycoform ratio,Citation15 as well as in stability to chaotropes.Citation16 However, techniques for directly linking protein conformational features with disease phenotypes are lacking, and conventional immunohistological staining of prion aggregates in brain tissue sections cannot be utilized to distinguish PrP deposits associated with distinct prion strains.
Luminescent conjugated poly- and oligothiophenes (LCPs and LCOs) have been used as conformation-induced fluorescence emission reporters to distinguish between specific protein aggregates.Citation17-25 The anionic LCP, polythiophene acetic acid (PTAA, ), also allowed for fluorescent spectral discrimination of immunohistochemically indistinguishable PrP aggregates in brain tissue sections from mice infected with different prion strains, as the emission profile from PTAA bound to PrP aggregates was shown to be strain dependent.Citation18,26,27 The aggregate-specific emission is achieved due to conformational restriction of the thiophene backbone upon interaction with specific protein aggregates. This restriction induces either a distinct twist or planarization of the backbone as well as potential stacking between adjacent polythiophene chains that can be observed as a unique strain-dependent emission profile from the dye.Citation18,26,27
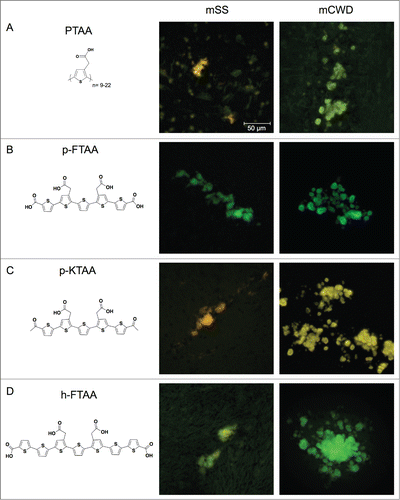
Figure 1. Chemical structures of thiophene based amyloid ligands and fluorescence images of stained PrP deposits in brain tissue sections from mice infected with mSS or mCWD. (A) PTAA, a polydisperse polythiophene. (B) p-FTAA, a chemically defined pentameric oligothiophene. (C) p-KTAA, a chemically defined pentameric oligothiophene. (D) h-FTAA, a chemically defined heptameric oligothiophene. Scale bar represents 50 μm.
In addition to the unique emission profiles, the altered conformation dependent optical transitions can also be observed from the excitation profiles or the fluorescence decay from the LCPs and LCOs. For the latter, fluorescence lifetime imaging (FLIM) proves to be an exceedingly sensitive method, as it is able to monitor distinct rates of fluorescence decay after excitation.Citation28,29 The decay rates do not depend on the concentration of the chromophore and FLIM is a direct approach for evaluating energy transfer between an excited molecule and its environment. In addition, LCOs bound to amyloid deposits exhibit unique excitation spectra that reflect the conformational constraints of the dyes upon interaction with the deposits and excitation profiles can be utilized to assess conformational alterations of the dyes.Citation20,22,30 Herein, we utilized the more sensitive methods of lifetime measurements and excitation spectroscopy, as well as emission spectroscopy from LCPs and LCOs bound to PrP deposits in brain tissue sections from mice inoculated with 2 distinct prion strains. The obtained results clearly demonstrate how multimodal fluorescence microscopy can be utilized for enhanced optical separation of PrP deposits associated with distinct prion strains. Furthermore, our data support the theory that changes in fluorescence are due to distinct conformational restriction of the thiophene backbone upon interaction with distinct supramolecular PrP structures.
Results
Emission profiles from LCP and LCO stained PrP deposits
PTAA, a polydisperse anionic LCP, () has been reported to provide distinct emission spectra for prion aggregates associated with 2 mouse adapted prion strains denoted murine chronic wasting diseases (mCWD) and murine sheep scrapie (mSS).Citation18 Frozen brain sections from mice infected with these 2 strains were utilized for optical evaluation of PTAA and 3 chemically defined LCOs. The structurally related LCOs, p-FTAA (), p-KTAA (), and h-FTAA () were included in the study, since the length of the thiophene backbone as well as the position of the anionic groups along the backbone, have been identified as important structural determinants for thiophene-based ligands efficient for spectral separation of protein aggregates.Citation20,22,30 All 4 dyes successfully stained PrP aggregates associated with each of the distinct strains and the deposits were easily identifiable due to the bright fluorescence from the thiophene-based ligands (). For PTAA, some background staining was observed, whereas all of the chemically defined LCOs showed superior specific staining of the protein aggregates with low background from surrounding tissue.
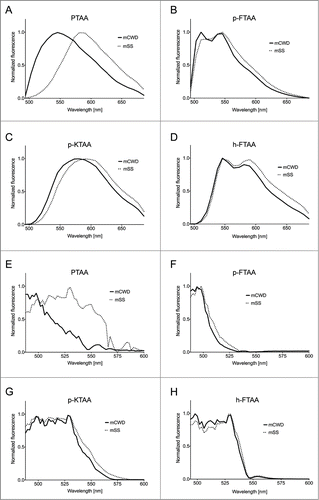
Evaluation of the LCP and LCO emission spectra showed that to differing degrees, each of the ligands were able to spectrally distinguish between mSS and mCWD plaques with PTAA providing the largest spectral differentiation (). In agreement with previous studies,Citation18,27 PTAA bound to mCWD deposits emitted light with an emission maximum around 550 nm, whereas PTAA stained mSS deposits displayed red-shifted emission spectra with a maximum intensity around 590 nm (). p-FTAA and h-FTAA displayed double-peaks in the emission spectra when bound to the PrP deposits, and mSS deposits displayed spectra with an increased intensity of the most red-shifted emission peak as well as a shoulder at longer wavelengths compared with the spectra from mCWD deposits stained by the corresponding LCO (). Similar to PTAA, a red-shift of the emission maxima toward longer wavelengths was observed from p-KTAA-stained mSS deposits compared with mCWD deposits stained by p-KTAA (). In contrast to p-FTAA and h-FTAA, the emission spectra from p-KTAA bound to PrP deposits lacked the characteristic double-peaks previously observed from a variety of LCOs bound to protein deposits.Citation20,22,30 Overall, all the dyes exhibited altered emission properties associated with pronounced planarization of the backbone and/or stacking between adjacent thiophene chains when bound to mSS deposits.
Figure 2. Emission and excitation spectra of thiophene based amyloid ligands bound to mSS (dotted line) or mCWD (black line) deposits. Emission spectra for (A) PTAA, (B) p-FTAA, (C) p-KTAA, and (D) h-FTAA. The emission spectra were recorded using excitation at 488 nm. Excitation spectra for (E) PTAA, (F) p-FTAA, (G) p-KTAA, and (H) h-FTAA. The excitation spectra were recorded having the emission locked for each probe and prion strain within the respective emission window.
Excitation profiles from LCP and LCO stained PrP deposits
In order to investigate the conformation-dependent interactions for each respective strain with an additional fluorescent mode, excitation spectra for each of the dyes bound to mCWD or mSS deposits samples were acquired. Using a tunable laser, we were able to acquire excitation spectra of each of the samples between 488 nm and 600 nm. Assessment of the excitation spectra showed that some dyes displayed strain-specific alteration of the excitation spectra (). For the polydisperse PTAA, an evident difference between the excitation spectra for the dye bound to mCWD vs. mSS deposits was observed (). PTAA bound to mSS deposits exhibited an excitation spectrum having a maximum around 535 nm and a shoulder around 560 nm, whereas a blue-shifted excitation spectrum was observed for PTAA bound to mCWD deposits. The strain-dependent shifts displayed in both the excitation- and emission spectra for PTAA, indicates that PTAA is more planar in the ground state as well as in the excited state when bound to mSS deposits.
In a similar fashion as PTAA, all the chemically defined LCOs bound to mSS deposits also displayed a red-shifted excitation spectrum with an evident shoulder at longer wavelengths compared with mCWD deposits stained by the corresponding ligand (). The shift was not as prominent as that observed for PTAA, suggesting that, in contrast to PTAA, the chemically defined LCOs may adopt rather similar conformations when bound to mCWD or mSS deposits. However, previous studiesCitation20,22,30 have shown that LCOs bound to recombinant Aβ1–42 fibrils display excitation maxima in a more blue-shifted region, 450 nm to 500 nm, and due to limitation of the tunable laser, this region cannot be recorded. Thus, the LCOs bound to mCWD or mSS deposits might display more pronounced differences in the excitation spectra at shorter wavelengths than 490 nm. The shoulder at longer wavelengths observed for LCOs bound to mSS deposits might be associated with increased π-stacking between adjacent thiophene backbones.Citation31-33 In general, the results from the excitation measurement confirmed that thiophene-based ligands adopt alternative conformations when bound to mSS or mCWD deposits and analogous to the emission measurements, PTAA was the most efficient ligand differentiating the deposits using excitation spectra in the accessible spectral range.
Cluster analysis of emission and excitation profiles
The strain-dependent spectral variations have previously been assessed by plotting the ratio of the emission intensity at distinct wavelengths.Citation18 However, a complete analysis of the entire spectral profile instead of the emission intensity at distinct wavelengths would provide more comprehensive information regarding the spectral variation from deposits within one strain, as well as spectral variations between deposits from different strains. Therefore, the emission- and excitation profiles were analyzed by hierarchical clustering based on Euclidean distance.Citation34 Spectra were collected from 50 different regions from protein deposits associated with the respective prion strain (Fig. S1). Thus, a total of 100 spectra for each probe were used. Each spectrum was rendered as a vector and the Euclidean distance was calculated as the square distance between 2 vectors. The distance was calculated between vectors within mSS and mCWD as well as between the 2 strains (). The Euclidean distances were also used to build a hierarchy of clusters, showing the relationship between the individual vectors and merging vector clusters based on similarity.
Table 1. Mean Euclidean distances for the emission and excitation profiles within and between the prion strains
As shown in and , a comparison of the Euclidean distances can be presented as heat maps and dendrograms where spectra coming from the same distribution, mSS or mCWD deposits, should end up in the same cluster if the intra-strain variability is low and inter-strain variability is high. Each box in the heat map corresponds to the Euclidean distance between 2 spectra and the red-to-blue scale assigns the compared spectra with a color indicating vector similarity and clustering. Dark blue indicates a Euclidean distance of 0 (identical spectra) and dark red indicates a large distance (relative to the data set). The results of the hierarchial clustering of spectra based on Euclidean distance, visualized as heat maps and dendograms ( and ), clearly showed that mCWD and mSS deposits can be separated based on their spectra. For PTAA, both the emission- and excitation profiles were separated in 2 distinct clusters corresponding to mCWD or mSS deposits (A and ). Comparison of Euclidean distances for emission- and excitation profiles from PTAA-stained deposits from an individual strain resulted in a blue color, whereas a red color was obtained when comparing emission- and excitation profiles from deposits from opposite strains. Thus, the largest Euclidean distances were obtained between strains, verifying that the PTAA spectral profiles were homogeneous for individual strains, but considerably dissimilar for different strains ( and ).
Figure 3. Heat maps and dendrograms for Euclidean distances between pairs of emission spectra from 50 different regions from PrP deposits associated with the respective prion strain. Rows and columns in the heat maps are ordered according to hierarchical clustering of the spectra for (A) PTAA, (B) p-FTAA, (C) p-KTAA, and (D) h-FTAA. The lines above and left to the heat maps indicate if a specific spectrum is associated with mCWD (green) or mSS (orange). For all the dyes the emission profiles are separated into 2 distinct clusters corresponding to mCWD (green) or mSS (orange) deposits.
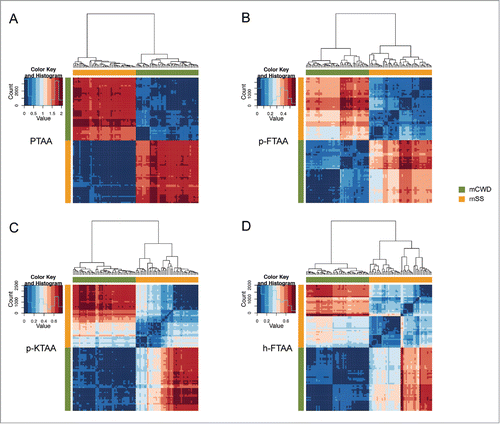
Figure 4. Heat maps and dendrograms for Euclidean distances between pairs of excitation spectra from 50 different regions from protein deposits associated with the respective prion strain. Rows and columns in the heat maps are ordered according to hierarchical clustering of the spectra for (A) PTAA, (B) p-FTAA, (C) p-KTAA, and (D) h-FTAA. The lines above and left to the heat maps indicate if a specific spectrum is associated with mCWD (green) or mSS (orange). For all the dyes the excitation profiles are separated into 2 distinct clusters corresponding to mCWD (green) or mSS (orange) deposits. For h-FTAA (D) the excitation profiles displayed some partial overlap.
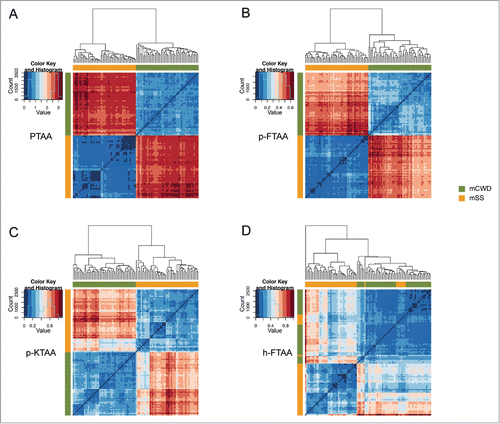
Analogous to PTAA, when analyzing the emission profiles for all the chemically defined LCOs, the emission data were separated into 2 clusters correlating to mSS and mCWD deposits (). As expected, the difference in the mean Euclidean distances between the prion strains was not as striking as for PTAA (). Nonetheless, for p-FTAA and p-KTAA, Euclidean distances for the excitation spectra also displayed a strain-specific difference (). In contrast, the excitation profiles from h-FTAA bound to mSS or mCWD displayed partial overlap (). Overall, the cluster analysis revealed that PTAA was the most efficient dye in separating mSS and mCWD deposits by emission- and excitation profiles. The chemically defined LCOs, p-FTAA, and p-KTAA also displayed a clear strain-specific differentiation when using both the fluorescent modalities. For the heptameric LCO, h-FTAA, clear strain-specific differentiation could only be obtained using emission profiles.
Fluorescence lifetime imaging of LCP and LCO stained PrP deposits
Finally, fluorescence lifetime imaging microscopy (FLIM) was employed to determine both differential states of binding and to observe minute variations in ligand-aggregate interactions. In contrast to the emission- and excitation measurements, the chemically defined LCOs displayed the most efficient separation of the strain specific prion deposits using FLIM. For PTAA, a narrow distribution of fluorescent decays (400 ps to 600 ps) was obtained from the dye bound to mSS deposits, whereas PTAA bound to CWD displayed a broader range of decays ranging from 300 ps to 800 ps (). Thus, the lifetime measurements from PTAA revealed an extensive overlap for the decays obtained from mSS and mCWD deposits. The broad distribution of fluorescence decays is most likely a consequence of the polydisperse nature of PTAA, since a mixture of thiophene chains with different lengths will exhibit different binding modes to the aggregates as well as give rise to a more extensive variation in energy transfer between this mixture of excited thiophene molecules and their environments.
Figure 5. Fluorescence lifetime imaging of thiophene based amyloid ligands bound to PrP deposits. Lifetime decay curves of (A) PTAA, (B) p-FTAA, (C) p-KTAA, and (D) h-FTAA bound to mCWD (black line) and mSS (dotted line) deposits. (E) Fluorescence lifetime images of LCP and LCO stained mCWD and mSS deposits in brain tissue section. The color bars represent lifetimes from 300 ps (orange) to 1100 ps (blue) and the images are color coded according to the representative lifetime. The fluorescence lifetimes were collected with excitation at 490 nm.
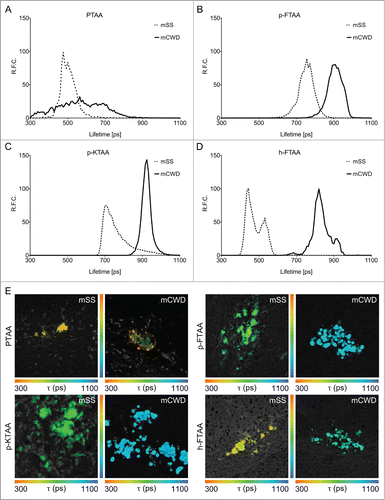
Comparison of the FLIM data from the chemically defined LCOs also supported the notion that LCO homogeneity leads to a narrower distribution of lifetimes. When bound to mCWD deposits, p-FTAA showed a narrow distribution of fluorescence decays between 800 ps to 1000 ps, whereas p-FTAA stained mSS deposits exhibited shorter decays (650 ps to 850 ps) (). p-KTAA displayed similar behavior to p-FTAA, having shorter fluorescence decays when bound to mSS deposits (). Although, there was some partial overlap between the fluorescence decays obtained from the strain-specific deposits, both p-FTAA and p-KTAA could be applied for efficient separation of the strain-specific PrP deposits by FLIM (). An enhanced separation of the PrP deposits could be achieved by h-FTAA. h-FTAA-stained mSS deposits revealed fluorescence decays between 400 ps to 600 ps, whereas the decays from h-FTAA bound to mCWD were considerably longer, 750 ps to 950 ps (). Hence, a complete separation of the fluorescence decays from the strain-specific deposits was observed for h-FTAA.
The difference in lifetime decays between the strain-specific deposits can also be visualized by color-coded fluorescence lifetime images (). All the chemically defined LCOs, displayed a strain-specific color difference and the color was rather uniform for the deposits associated with a distinct strain. The strain dependent difference in fluorescence decays for the LCOs was also confirmed by statistical analysis using empirical cumulative distribution functions (ecdfs) and Kolmogorov-Smirnov statistics (Fig. S2 Table S1).Citation35,36 The ecdf representations of the lifetimes for p-FTAA, p-KTAA, and h-FTAA showed a pronounced difference between the strains, while the data for the 2 strains stained with PTAA tend to overlap (Fig. S2). The LCOs also displayed larger D values (maximum vertical difference between the ecdfs) between strains than D values within the strain, verifying that intra-strain variability was low and inter-strain variability was high (Table S1). In contrast, for PTAA, several D values within the strains were larger than between the strains. Overall, the FLIM experiments revealed that the chemically defined LCOs were the most efficient dyes in separating mSS and mCWD deposits with fluorescence decay distributions.
Discussion
Currently, efficient techniques for assessing distinct conformational features of protein aggregates are limited. However, the development of such techniques is essential as the presence of multiple aggregated conformations can be found in a variety neurodegenerative proteinopathies. In analogy to the prion strain phenomenon, distinct aggregated conformers might give rise to specific disease phenotypes.Citation8-12 Herein we showed that histological staining with thiophene-based amyloid ligands in combination with multimodal fluorescence microscopy offer a straight forward methodology to assign strain-specific prion deposits. In addition, through the use of homogenous well-defined oligiothiophenes and multiple fluorescent techniques, we were able to show that the observed alterations in fluorescence are due to distinct conformational restriction of the thiophene backbone upon interaction with PrP aggregates associated with distinct prion strains. These results strongly support the notion of conformational variations in the tertiary and/or quaternary structure of the PrP aggregates as a determinant of prion strains. Alternatively, the differences in fluorescence characteristics of oligothiopenes might arise due different chemical composition of the PrP aggregates. Many studiesCitation37,38 have shown that PrPSc forms tight complexes and co-purifies with other molecules, such as glycosaminoglycans, cholesterol, lipids, or polysaccharide scaffolds, and these components might influence the aggregation process of PrPCitation39 and potentially the binding of LCOs and LCPs to PrP deposits.
LCPs and LCOs bind preferentially to protein aggregates with repetitive cross β-sheet and recent studies have also shown that minor alterations of the chemical composition of the thiophene-based dye influence the dyes capability to distinguish different protein aggregate topologies.Citation20,22,30 In the present study, all the LCPs and LCOs included in the study could indeed be utilized as fluorescent ligands for optical separation of prion strain-specific PrP deposits. First, when using emission- and excitation profiles, PTAA was identified as the most effective compound, whereas p-FTAA, p-KTAA, and h-FTAA showed less pronounced strain-associated spectral alterations. Compared with the chemically defined pentameric and heptameric LCOs, PTAA has a polydisperse composition with longer thiophene backbones consisting of 9 to 22 thiophene units. Hence, the enhanced spectral alterations observed for PTAA can be a direct consequence of the length of the thiophene backbone. For PTAA a distinct strain-dependent difference in both the emission- and excitation maximum was observed, suggesting that there are conformational differences in both the ground state and the excited state of the molecule when bound to mCWD or mSS deposits. Clearly, the conjugated backbone of PTAA is more planar when bound to mSS deposits. This planarization was not as striking for the LCOs, as LCO-stained mSS deposits only displayed a minor red-shift in the emission profiles compared with LCO-stained mCWD deposits. Hence, LCOs might lack the ability to undergo the same prominent conformational alteration as PTAA, implicating that a certain length of the thiophene backbone is necessary to achieve an efficient strain-dependent planarization of the backbone. In addition, for PTAA all the thiophene units are functionalized with an acetic acid moiety and the relative positioning of these side chains will also influence the conformational freedom of the dye as well as the electrostatic interactions between the deposits and the dye. The latter is highly relevant, as it was recently shown that efficient binding of the most conventionally used amyloid ligand, Congo Red, to amyloid fibrils is highly dependent on electrostatic interactions and hydrogen bonding.Citation40 However, to investigate these structure-activity relationships in a more refined manner, a library consisting of chemically defined LCOs with longer thiophene backbones and alternative positioning of acetic acid motifs are required. From the results reported herein, we conclude that the chemical composition of the thiophene-based dye impacts its capability for spectral separation of different PrP aggregate topologies.
In earlier studies, correlation diagrams displaying the ratio of the emission intensity at specific wavelengths have been utilized to compare strain-dependent spectral variations of PTAA-stained deposits.Citation18,26,27 Herein, the complete emission- and excitation profiles were analyzed by hierarchical clustering based on Euclidean distance. First, the cluster analysis could be utilized to certify that spectral profiles were considerably different for PrP deposits associated with individual prion strains. This form of analysis allows for comparison of considerable amounts of spectra collected from distinct protein deposits to assess if there is a difference between specific populations of protein aggregates. Second, the analysis also offered the possibility to investigate the homogeneity or heterogeneity within a population of protein aggregates and herein we found that the spectral profiles were rather homogeneous for the individual strains. Altogether, this complete analysis of the spectral profile provided comprehensive information regarding the spectral variation from deposits within one strain, as well as spectral variations between deposits from different strains. As multiple aggregated protein conformations have been observed in sporadic Creutzfeldt–Jakob disease (sCJD)Citation14,Citation41-43 as well as in AD, synucleinopathies and tauopathies,Citation8-12 we foresee that this cluster analysis in combination with LCP and LCO staining will be valuable for exploring heterogenic populations of protein deposits for a variety of neurodegenerative proteinopathies. In addition, the methodology can also be applied for studying other phenomena, such as strain adaption and strain competitionCitation27 or time-dependent aggregate maturation.Citation24
Since conformational alterations of the thiophene backbone can be observed as distinct emission- and excitation profiles, we anticipated that these alterations should also be observed in the fluorescence decays from the LCPs and LCOs. This assumption was verified in the set of experiments utilizing FLIM. All 4 thiophene-based dyes display strain-dependent fluorescent decays. In addition, the FLIM data also provide some crucial information regarding the underlying conformational differences between LCOs bound to mSS or mCWD deposits. When stained by LCOs, mSS deposits displayed shorter decays compared with mCWD. These shorter decay times might be associated with increased π-stacking between adjacent thiophene backbones,Citation32,33 suggesting that the LCOs binds in a different fashion to mSS deposits than to mCWD deposits. In the excitation profiles, a shoulder at longer wavelengths was also observed for LCOs bound to mSS deposits when compared with LCO stained mCWD deposits. Such a shoulder at longer wavelengths is also indicative of π-stacking between adjacent thiophene backbones.Citation31,32 Thus, increased π-stacking between adjacent thiophene backbones is probably the major conformational alteration underlying the observed strain-dependent optical properties from the LCOs. In contrast to the LCOs, PTAA displayed broader and overlapping fluorescent decays, indicating that the polydisperse nature of PTAA gives rise to a range of different fluorescent decay times. Compared with emission- and excitation spectroscopy, FLIM is a more sensitive method, as it measures all energy transfer between an excited molecule and its environment independent of the concentration of the molecule. The heterogeneity in chain-length for PTAA is presumably not optimal for FLIM. It was also particularly interesting that the longest LCO, h-FTAA, containing 7 thiophene units and 2 carboxylic moieties flanking the conjugated backbone, provided an enhanced prion strain discrimination by fluorescence decay separation compared with the pentameric LCOs. This result clearly suggests that there is room for additional chemical improvements in structural design for achieving optimal thiophene-based probes for FLIM.
The findings described above verify that PTAA can be utilized for spectral assignment of prion deposits associated with distinct strains.Citation18,26,27 In addition, the toolbox of ligands for strain-dependent assessment of prion deposits can be expanded with chemically defined LCOs that can also be assessed using the more sensitive method of FLIM. In contrast to PTAA, the chemically defined LCOs can also be utilized for optical non-invasive in vivo imaging of protein aggregates by multi-photon spectroscopy.Citation20,44 As FLIM can also be applied for non-invasive imaging,Citation45 the LCOs might potentially be applied for strain-dependent assessment of prion deposits in vivo, as well as for kinetic studies of the strain-dependent protein aggregation events in real-time. Such studies could also be realistic for a variety of neurodegenerative proteinopathies as well as other amyloidoses.
Chemically defined LCOs will also be essential for developing the methodology toward other technical platforms than conventional fluorescence microscopy. Due to the diffraction limited resolution of optical microscopes, the detection of smaller protein aggregates (<0.25 μm) is restricted. Thus, the technique presented herein might not be sensitive enough for detection of PrP deposits associated with some prion strains or smaller aggregated species occurring during earlier stages of the protein aggregation process. To overcome this limitation, of optical microscopy, superresolution techniques, including stimulated emission depletion (STED)Citation46 and binding-activated localization microscopy (BALM),Citation47 that provide up to 10-fold higher resolution have been developed and it was recently shown that p-FTAA could be utilized for BALM imaging of α-synuclein fibrils.Citation48 Hence, the spectral shifts observed in the emission- and excitation profiles from LCOs bound to distinct protein deposits could potentially be recorded by BALM using multiple spectral channels and such a set up might offer the exciting possibility to visualize different aggregated conformations with higher resolution.
In conclusion, we have presented a multimodal fluorescence based methodology for the assignment of prion strain specific PrP deposits stained by thiophene based ligands. In addition, our findings verified that alterations in fluorescence are due to distinct conformational restriction of the thiophene backbone upon interaction with prion aggregates associated with distinct prion strains. We foresee that the methodology described herein will expand the toolbox of techniques for detecting structural differences among discrete protein aggregates, as well as provide novel insights regarding the interplay between protein conformational features and disease phenotypes for a variety of neurodegenerative proteinopathies.
Materials and Methods
LCP/LCO synthesis and staining of brain tissue cryosections
The syntheses of PTAA, p-FTAA, p-KTAA, and h-FTAA have been reported elsewhere.Citation20,22,49,50 All the probes were dissolved in deionized water to a final concentration of 1.5 mM and all the histological staining of the prion infected material was performed in a P3 facility. Brain tissue cryosections were dried for 1 h and fixed in 100% ethanol for 10 min followed by 70% ethanol for 5 min. After washing in deionized water, the sections were equilibrated in incubation buffer solution, 100 mM sodium carbonate, pH 10.2 (PTAA), or 100 mM phosphate buffer saline (PBS), pH 7.4 (p-FTAA, h-FTAA, and p-KTAA). The probes were diluted in incubation buffer to a final concentration of 3 μM, added to the brain sections and incubated for 30 min in room temperature. After washing in incubation buffer the slides were mounted with mounting media (DAKO). The mounting medium was allowed to solidify over night before the rims were sealed with nail polish and prior to analysis, the slides was decontaminated with 2 M NaOH.
Fluorescence microscopy emission and excitation spectra
The spectra were recorded with a LSM 780 Zeiss confocal laser scanning microscope equipped with a 32 channel QUASAR GaAsP spectral array detector and a tunable In Tune laser (488–640 nm). In emission mode, all probes were excited with a laser tuned to 488 nm and the emitted light was detected in steps of 8.7 nm from 488–686 nm. Excitation measurements were taken in steps of 2 nm from 488–600 nm with the emission locked for each probe and prion strain within the respective emission window. Spectra were collected from 10 individual spots within 5 plaques for each probe and strain.
Fluorescence lifetime imaging (FLIM)
Fluorescence lifetime images were acquired using an inverted Zeiss (Axio Observer.Z1) LSM 780 microscope (Carl Zeiss MicroImaging GmbH) equipped with a modular FLIM system from Becker and Hickl. In this setup the emitted photons were routed through the Direct coupling (DC) confocal port of the Zeiss LSM 780 scanning unit and detected by a Becker and Hickl HPM-100-40 hybrid detector. Data were recorded by a Becker and Hickl Simple-Tau 152 system (SPC-150 TCSPC FLIM module) with the instrument recording software SPCM version 9.42 in the FIFO image mode, 256 × 256 pixels, using 256 time channels (Becker and Hickl GmbH). For all acquisitions, a T80R20 main beam splitter was used and the pinhole was set to 20.2 μm. Scanning area was set to 235.7 μm × 235.7 μm, with a scanning resolution of 512 × 512 pixels. Further a Plan-Apochromat 40 × /1.3 Oil DIC objective lens was used and a 510 nm longpass filter was positioned in front of the hybrid PMT. Excitation utilized the 490 nm laser line from the pulsed tunable In Tune laser (Carl Zeiss MicroImaging GmbH) with a repetition rate of 40 MHz. Data was subsequently analyzed in SPCImage version 3.9.4 (Becker and Hickl GmbH) fitting each of the acquired decay curves to a tri-exponential function and color coded images showing the intensity-weighted mean lifetimes were generated with the same software.
Statistical analysis
The software R was used in all the statistical analyzes and all the data was normalized before any statistical calculations.Citation51 The emission and excitation data were analyzed by hierarchical clustering based on Euclidean distance. Each spectrum was rendered as a vector and the Euclidean distance was calculated as the square distance between 2 vectors. The Euclidean distance was calculated between vectors within mSS and mCWD as well as between the 2 strains. These Euclidean distances were used to build a hierarchy of clusters, showing relations between individual spectra and merging clusters of spectra based on similarity. The clusters are presented in dendrograms where the height of the branch in vertical dimension is proportional to the distance between the connected clusters. The Euclidean distances between all pairs of spectra are also presented in heat maps, where the color in each box in the matrix describes the Eucleadian distance between 2 spectra. Spectra were collected from 10 individual areas within 5 plaques for each probe and strain. Thus, 100 spectra for each probe were used and the algorithm distributes these hundred spectra according to similarity.
In order to analyze the difference between the strains by using FLIM, empirical cumulative distribution functions (ecdfs) corresponding to the lifetime distributions were calculated. The lifetime ecdf represents the probability of obtaining a given lifetime. Kolmogorov-Smirnov statistics were used to calculate the maximum vertical difference, D, between the ecdfs.Citation52 Data sets from the same distribution should have a low D value, while data from different data sets should have a high value with 1 as maximum. Lifetimes were measured on 3 different plaques for each probe and strain, which means that 6 lifetime distributions were analyzed for each probe.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental_Material.pdf
Download PDF (228.4 KB)Funding
Our work is supported by the Swedish Foundation for Strategic Research (KPR Nilsson, R Simon, K Magnusson), the Swedish Research Council (P Hammarström), and the Linköping University Center for Neuroscience (D.S.). KPR Nilsson is financed by an ERC Starting Independent Researcher Grant (Project: MUMID) from the European Research Council. R Simon and K Magnusson are enrolled in the doctoral program Forum Scientum. Support by BILS (Bioinformatics Infrastructure for Life Sciences), M Larsson, and E Freyhult are gratefully acknowledged.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- Prusiner SB. Novel proteinaceous infectious particles cause scrapie. Science 1982 216:136-44 PMID:6801762 http://dx.doi.org/10.1126/science.6801762
- Prusiner SB. Molecular biology of prion diseases. Science 1991 252:1515-22 PMID:1675487 http://dx.doi.org/10.1126/science.1675487
- Collinge J, Clarke AR. A general model of prion strains and their pathogenicity. Science 2007 318:930-6 PMID:17991853 http://dx.doi.org/10.1126/science.1138718
- Fraser H, Dickinson AG. The sequential development of the brain lesion of scrapie in three strains of mice. J Comp Pathol 1968 78:301-11 PMID:4970192 http://dx.doi.org/10.1016/0021-9975(68)90006-6
- Fraser H, Dickinson AG. Scrapie in mice. Agent-strain differences in the distribution and intensity of grey matter vacuolation. J Comp Pathol 1973 83:29-40 PMID:4199908 http://dx.doi.org/10.1016/0021-9975(73)90024-8
- Bruce ME, McBride PA, Farquhar CF. Precise targeting of the pathology of the sialoglycoprotein, PrP, and vacuolar degeneration in mouse scrapie. Neurosci Lett 1989 102:1-6 PMID:2550852 http://dx.doi.org/10.1016/0304-3940(89)90298-X
- Aguzzi A. Understanding the diversity of prions. Nat Cell Biol 2004 6:290-2 PMID:15057242 http://dx.doi.org/10.1038/ncb0404-290
- Clavaguera F, Lavenir I, Falcon B, Frank S, Goedert M, Tolnay M. “Prion-like” templated misfolding in tauopathies. Brain Pathol 2013 23:342-9 PMID:23587140 http://dx.doi.org/10.1111/bpa.12044
- Clavaguera F, Akatsu H, Fraser G, Crowther RA, Frank S, Hench J, Probst A, Winkler DT, Reichwald J, Staufenbiel M, et al. Brain homogenates from human tauopathies induce tau inclusions in mouse brain. Proc Natl Acad Sci U S A 2013 110:9535-40 PMID:23690619 http://dx.doi.org/10.1073/pnas.1301175110
- Lu JX, Qiang W, Yau WM, Schwieters CD, Meredith SC, Tycko R. Molecular structure of β-amyloid fibrils in Alzheimer's disease brain tissue. Cell 2013 154:1257-68 PMID:24034249 http://dx.doi.org/10.1016/j.cell.2013.08.035
- Guo JL, Covell DJ, Daniels JP, Iba M, Stieber A, Zhang B, Riddle DM, Kwong LK, Xu Y, Trojanowski JQ, et al. Distinct α-synuclein strains differentially promote tau inclusions in neurons. Cell 2013 154:103-17 PMID:23827677 http://dx.doi.org/10.1016/j.cell.2013.05.057
- Bousset L, Pieri L, Ruiz-Arlandis G, Gath J, Jensen PH, Habenstein B, Madiona K, Olieric V, Böckmann A, Meier BH, et al. Structural and functional characterization of two alpha-synuclein strains. Nat Commun 2013 4:2575 PMID:24108358 http://dx.doi.org/10.1038/ncomms3575
- Bessen RA, Marsh RF. Biochemical and physical properties of the prion protein from two strains of the transmissible mink encephalopathy agent. J Virol 1992 66:2096-101 PMID:1347795
- Polymenidou M, Stoeck K, Glatzel M, Vey M, Bellon A, Aguzzi A. Coexistence of multiple PrPSc types in individuals with Creutzfeldt-Jakob disease. Lancet Neurol 2005 4:805-14 PMID:16297838 http://dx.doi.org/10.1016/S1474-4422(05)70225-8
- Hill AF, Joiner S, Beck JA, Campbell TA, Dickinson A, Poulter M, Wadsworth JD, Collinge J. Distinct glycoform ratios of protease resistant prion protein associated with PRNP point mutations. Brain 2006 129:676-85 PMID:16415305 http://dx.doi.org/10.1093/brain/awl013
- Peretz D, Williamson RA, Legname G, Matsunaga Y, Vergara J, Burton DR, DeArmond SJ, Prusiner SB, Scott MR. A change in the conformation of prions accompanies the emergence of a new prion strain. Neuron 2002 34:921-32 PMID:12086640 http://dx.doi.org/10.1016/S0896-6273(02)00726-2
- Nilsson KPR, Aslund A, Berg I, Nyström S, Konradsson P, Herland A, Inganäs O, Stabo-Eeg F, Lindgren M, Westermark GT, et al. Imaging distinct conformational states of amyloid-beta fibrils in Alzheimer's disease using novel luminescent probes. ACS Chem Biol 2007 2:553-60 PMID:17672509 http://dx.doi.org/10.1021/cb700116u
- Sigurdson CJ, Nilsson KPR, Hornemann S, Manco G, Polymenidou M, Schwarz P, Leclerc M, Hammarström P, Wüthrich K, Aguzzi A. Prion strain discrimination using luminescent conjugated polymers. Nat Methods 2007 4:1023-30 PMID:18026110 http://dx.doi.org/10.1038/nmeth1131
- Philipson O, Hammarström P, Nilsson KPR, Portelius E, Olofsson T, Ingelsson M, Hyman BT, Blennow K, Lannfelt L, Kalimo H, et al. A highly insoluble state of Abeta similar to that of Alzheimer's disease brain is found in Arctic APP transgenic mice. Neurobiol Aging 2009 30:1393-405 PMID:18192084 http://dx.doi.org/10.1016/j.neurobiolaging.2007.11.022
- Åslund A, Sigurdson CJ, Klingstedt T, Grathwohl S, Bolmont T, Dickstein DL, Glimsdal E, Prokop S, Lindgren M, Konradsson P, et al. Novel pentameric thiophene derivatives for in vitro and in vivo optical imaging of a plethora of protein aggregates in cerebral amyloidoses. ACS Chem Biol 2009 4:673-84 PMID:19624097 http://dx.doi.org/10.1021/cb900112v
- Nilsson KPR, Ikenberg K, Åslund A, Fransson S, Konradsson P, Röcken C, Moch H, Aguzzi A. Structural typing of systemic amyloidoses by luminescent-conjugated polymer spectroscopy. Am J Pathol 2010 176:563-74 PMID:20035056 http://dx.doi.org/10.2353/ajpath.2010.080797
- Klingstedt T, Åslund A, Simon RA, Johansson LBG, Mason JJ, Nyström S, Hammarström P, Nilsson KPR. Synthesis of a library of oligothiophenes and their utilization as fluorescent ligands for spectral assignment of protein aggregates. Org Biomol Chem 2011 9:8356-70 PMID:22051883 http://dx.doi.org/10.1039/c1ob05637a
- Heilbronner G, Eisele YS, Langer F, Kaeser SA, Novotny R, Nagarathinam A, Åslund A, Hammarström P, Nilsson KPR, Jucker M. Seeded strain-like transmission of β-amyloid morphotypes in APP transgenic mice. EMBO Rep 2013 14:1017-22 PMID:23999102 http://dx.doi.org/10.1038/embor.2013.137
- Nyström S, Psonka-Antonczyk KM, Ellingsen PG, Johansson LBG, Reitan N, Handrick S, Prokop S, Heppner FL, Wegenast-Braun BM, Jucker M, et al. Evidence for age-dependent in vivo conformational rearrangement within Aβ amyloid deposits. ACS Chem Biol 2013 8:1128-33 PMID:23521783 http://dx.doi.org/10.1021/cb4000376
- Bett C, Kurt TD, Lucero M, Trejo M, Rozemuller AJ, Kong Q, Nilsson KPR, Masliah E, Oldstone MB, Sigurdson CJ. Defining the conformational features of anchorless, poorly neuroinvasive prions. PLoS Pathog 2013 9:e1003280 PMID:23637596 http://dx.doi.org/10.1371/journal.ppat.1003280
- Sigurdson CJ, Nilsson KP, Hornemann S, Heikenwalder M, Manco G, Schwarz P, Ott D, Rülicke T, Liberski PP, Julius C, et al. De novo generation of a transmissible spongiform encephalopathy by mouse transgenesis. Proc Natl Acad Sci U S A 2009 106:304-9 PMID:19073920 http://dx.doi.org/10.1073/pnas.0810680105
- Nilsson KPR, Joshi-Barr S, Winson O, Sigurdson CJ. Prion strain interactions are highly selective. J Neurosci 2010 30:12094-102 PMID:20826672 http://dx.doi.org/10.1523/JNEUROSCI.2417-10.2010
- Wolfgang B, Advanced T-CSPCT. (Springer Series in Chemical Physics, Volume 81), Berlin Heidelberg: Springer 2005 ISBN: 9783540260479.
- Bugiel I, König K, Wabnitz H. Investigation of cells by fluorescence laser scanning microscopy with subnanosecond time resolution. Lasers Life Sciences 1989 3:1-7.
- Klingstedt T, Shirani H, Åslund KOA, Cairns NJ, Sigurdson CJ, Goedert M, Nilsson KPR. The structural basis for optimal performance of oligothiophene-based fluorescent amyloid ligands: conformational flexibility is essential for spectral assignment of a diversity of protein aggregates. Chemistry 2013 19:10179-92 PMID:23780508 http://dx.doi.org/10.1002/chem.201301463
- Miteva T, Palmer L, Kloppenburg L, Neher D, Bunz UHF. Interplay of thermochromicity and liquid crystalline behavior in poly(p-phenyleneethynylene)s π-π interactions or planarization of the conjugated backbone? Macromolecules 2000 33:652-4 http://dx.doi.org/10.1021/ma9912397
- Kim J, Swager TM. Control of conformational and interpolymer effects in conjugated polymers. Nature 2001 411:1030-4 PMID:11429599 http://dx.doi.org/10.1038/35082528
- Ma Y-Z, Shaw RW, Yu X, O’Neill HM, Hong K. Excited-state dynamics of water-soluble polythiophene derivatives: temperature and side-chain length effects. J Phys Chem B 2012 116:14451-60 PMID:23140246 http://dx.doi.org/10.1021/jp304526h
- Danielsson P-E. Euclidean distance mapping. Computer Graphics and Image Processing 1980 14:227-48 http://dx.doi.org/10.1016/0146-664X(80)90054-4
- Esposito A, Dohm CP, Kermer P, Bähr M, Wouters FS. alpha-Synuclein and its disease-related mutants interact differentially with the microtubule protein tau and associate with the actin cytoskeleton. Neurobiol Dis 2007 26:521-31 PMID:17408955 http://dx.doi.org/10.1016/j.nbd.2007.01.014
- Bhatta H, Goldys EM. Characterization of yeast strains by fluorescence lifetime imaging microscopy. FEMS Yeast Res 2008 8:81-7 PMID:18215225 http://dx.doi.org/10.1111/j.1567-1364.2007.00340.x
- Gabizon R, McKinley MP, Prusiner SB. Purified prion proteins and scrapie infectivity copartition into liposomes. Proc Natl Acad Sci U S A 1987 84:4017-21 PMID:3108886 http://dx.doi.org/10.1073/pnas.84.12.4017
- Shaked GM, Meiner Z, Avraham I, Taraboulos A, Gabizon R. Reconstitution of prion infectivity from solubilized protease-resistant PrP and nonprotein components of prion rods. J Biol Chem 2001 276:14324-8 PMID:11152454
- Vieira TCRG, Cordeiro Y, Caughey B, Silva JL. Heparin binding confers prion stability and impairs its aggregation. FASEB J 2014 http://dx.doi.org/10.1096/fj.13-246777, In press PMID:24648544
- Schütz AK, Soragni A, Hornemann S, Aguzzi A, Ernst M, Böckmann A, Meier BH. The amyloid-Congo red interface at atomic resolution. Angew Chem Int Ed Engl 2011 50:5956-60 PMID:21591034 http://dx.doi.org/10.1002/anie.201008276
- Puoti G, Giaccone G, Rossi G, Canciani B, Bugiani O, Tagliavini F. Sporadic Creutzfeldt-Jakob disease: co-occurrence of different types of PrP(Sc) in the same brain. Neurology 1999 53:2173-6 PMID:10599800 http://dx.doi.org/10.1212/WNL.53.9.2173
- Giles K, De Nicola GF, Patel S, Glidden DV, Korth C, Oehler A, DeArmond SJ, Prusiner SB. Identification of I137M and other mutations that modulate incubation periods for two human prion strains. J Virol 2012 86:6033-41 PMID:22438549 http://dx.doi.org/10.1128/JVI.07027-11
- Cali I, Castellani R, Alshekhlee A, Cohen Y, Blevins J, Yuan J, Langeveld JP, Parchi P, Safar JG, Zou WQ, et al. Co-existence of scrapie prion protein types 1 and 2 in sporadic Creutzfeldt-Jakob disease: its effect on the phenotype and prion-type characteristics. Brain 2009 132:2643-58 PMID:19734292 http://dx.doi.org/10.1093/brain/awp196
- Wegenast-Braun BM, Skodras A, Bayraktar G, Mahler J, Fritschi SK, Klingstedt T, Mason JJ, Hammarström P, Nilsson KPR, Liebig C, et al. Spectral discrimination of cerebral amyloid lesions after peripheral application of luminescent conjugated oligothiophenes. Am J Pathol 2012 181:1953-60 PMID:23041059 http://dx.doi.org/10.1016/j.ajpath.2012.08.031
- Abe K, Zhao L, Periasamy A, Intes X, Barroso M. Non-invasive in vivo imaging of near infrared-labeled transferrin in breast cancer cells and tumors using fluorescence lifetime FRET. PLoS One 2013 8:e80269 PMID:24278268 http://dx.doi.org/10.1371/journal.pone.0080269
- Klar TA, Jakobs S, Dyba M, Egner A, Hell SW. Fluorescence microscopy with diffraction resolution barrier broken by stimulated emission. Proc Natl Acad Sci U S A 2000 97:8206-10 PMID:10899992 http://dx.doi.org/10.1073/pnas.97.15.8206
- Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, Lippincott-Schwartz J, Hess HF. Imaging intracellular fluorescent proteins at nanometer resolution. Science 2006 313:1642-5 PMID:16902090 http://dx.doi.org/10.1126/science.1127344
- Ries J, Udayar V, Soragni A, Hornemann S, Nilsson KPR, Riek R, Hock C, Ewers H, Aguzzi AA, Rajendran L. Superresolution imaging of amyloid fibrils with binding-activated probes. ACS Chem Neurosci 2013 4:1057-61 PMID:23594172 http://dx.doi.org/10.1021/cn400091m
- Ding L, Jonforsen M, Roman LS, Andersson MR, Inganäs O. Photovoltaic cells with a conjugated polyelectrolyte. Synth Met 2000 110:133-40 http://dx.doi.org/10.1016/S0379-6779(99)00284-2
- Simon R, Shirani H, Åslund A, Bäck M, Haroutunian V, Gandy S, Nilsson KPR. Pentameric thiophene based ligands that spectrally discriminate disease-associated protein aggregates display distinct solvatochromism and viscosity induced spectral shifts. Chemistry 2014 (Forthcoming).
- R Core team. (2013) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL http://www.R-project.org/.
- Demidenko E. Kolmogorov-Smirnov Test for Image Comparison. Computational Science and Its Applications – ICCSA 2004, 2004 3046:933-9.
