Abstract
Molecular chaperones regulate essential steps in the propagation of yeast prions. Yeast prions possess domains enriched in glutamines and asparagines that act as templates to drive the assembly of native proteins into beta-sheet-rich, amyloid-like fibrils. Several recent studies highlight a significant and complex function for Hsp40 co-chaperones in propagation of prion elements in yeast. Hsp40 co-chaperones bind non-native polypeptides and transfer these clients to Hsp70s for refolding or degradation. How Hsp40 co-chaperones bind amyloid-like prion conformers that are enriched in hydrophilic residues such as glutamines and asparagines is a significant question in the field. Interestingly, selective recognition of amyloid-like conformers by distinct Hsp40s appears to confer opposing actions on prion assembly. For example, the Type I Hsp40 Ydj1 and Type II Hsp40 Sis1 bind different regions within the prion protein Rnq1 and function respectively to inhibit or promote [RNQ+] prion assembly. Thus, substrate selectivity enables distinct Hsp40s to act at unique steps in prion propagation.
Molecular Chaperones and Yeast Prions
Proteins adopt a diverse and dynamic array of structural conformations. Prions are unique in that these proteins induce conversion of the soluble, native structure into the prion conformer with a high propensity to self-assemble into beta-sheet-rich, amyloid-like fibrils.Citation1 Extensive investigation of prion biogenesis in the budding yeast Saccharomyces cerevisiae has uncovered some of the basic mechanisms underlying prion assembly into amyloid-like fibrils and inheritance of the prion state. One intriguing development in this story was the intimate role for heat shock protein (HSP) molecular chaperones in these pathways.Citation2,Citation3 Indeed, numerous yeast prions are dependent upon molecular chaperones for efficient maintenance and propagation of prion structures.Citation4,Citation5 On the other hand, overexpression of some molecular chaperones “cure” yeast of the heritable prion suggesting molecular chaperones antagonize prion assembly.Citation4,Citation6,Citation7 How such opposing activities efficiently coordinate prion assembly into amyloid-like fibrils and propagation of the prion state inside the cell is an outstanding question in the field. Study of this process is significant because amyloid-like fibrils accumulate in numerous conformational disorders.Citation8,Citation9 However, the connection between amyloid assembly and neuronal cell death is still controversial as several recent studies implicate the assembly of amyloid-like fibrils as benign or even protective.Citation10–Citation12 In addition, prions found in S. cerevisiae possess domains enriched in glutamines (Gln) and asparagines (Asn),Citation13 resembling proteins with expanded polyglutamine repeats (such as human huntingtin and several ataxins) that are very susceptible to aggregation.Citation14,Citation15 Many molecular chaperones are functionally conserved from yeast to humans, and as such, studying how molecular chaperones modulate prion propagation yields substantial mechanistic insight on the regulation of amyloid assembly in conformational disorders.
Several classes of molecular chaperone are implicated in prion propagation. For example, the AAA+ protein remodeling factor Hsp104 is required for propagation of several prions in yeast.Citation4,Citation16,Citation17 Hsp104 is proposed to shear prion polymers to generate “seeds” that drive conversion of native protein into the prion conformation.Citation18–Citation21 Hsp70 molecular chaperones also regulate prion propagation although the particular function depends on the Hsp70 class and specific yeast prion. For example, mutations in the Hsp70 Ssa1 destabilize [PSI+] prion propagation while overexpression of Hsp70s from the Ssa family can stabilize the [PSI+] state.Citation22–Citation24 Interestingly, overexpression of Ssa1 has been shown to cure yeast of the prion [URE3].Citation6,Citation7 In contrast, Hsp70s of the Ssb family appear to antagonize [PSI+] prion propagation.Citation24–Citation26 As a result, protein flux through Hsp70 refolding pathways is a crucial step in prion biogenesis. Hsp70 chaperone activity is tightly coordinated by Hsp40 co-chaperones (also known as J-proteins). Additionally, Hsp40 and Hsp70 chaperones cooperate with Hsp104 to refold aggregated proteins.Citation27 Several recent studies underscore a complex, yet fundamental role for Hsp40 co-chaperones in prion assembly and propagation.Citation5,Citation12,Citation28–Citation31 In this review we first describe the Hsp40 co-chaperone family and basic mechanisms underlying Hsp40:substrate recognition. Then, general roles for Hsp40s in propagation of the yeast prions [PSI+] and [URE3], and [RNQ+]/[PIN+] are discussed. Recent studies investigating the function of Hsp40s on assembly of the [RNQ+] prion are emphasized to highlight novel mechanisms in which Hsp40s bind Gln/Asn-rich prion proteins and modulate the accumulation of toxic or benign prion conformers. We propose that the distinct binding preferences of individual Hsp40s determine specific Hsp40 actions in prion assembly and propagation.
Protein Quality Control by Hsp40 Molecular Chaperones
Hsp40 co-chaperones are essential partners in Hsp70 function.Citation32 Hsp40s share a highly conserved region called a J-domain that stimulates the intrinsic ATPase activity of its partner Hsp70.Citation33 ATP hydrolysis causes a series of conformational changes that increase the affinity of client:Hsp70 interactions.Citation34,Citation35 Client release from Hsp70 is induced when ADP is replaced with ATP by a Hsp70 nucleotide exchange factor.Citation36 The Hsp40 J-domain alone appears sufficient to maintain basic cellular processes required for physiological growth.Citation37 However, based upon homology to the J-domain from the founder Hsp40 in Escherichia coli (DnaJ), there are twenty-two Hsp40s in budding yeast and forty-one Hsp40s in humans.Citation32,Citation38 Given the evolutionary expansion of the Hsp40 family, how these various Hsp40s specify Hsp70 function is an important unanswered question.
Importantly, Hsp40s utilize a variety of specialized domains outside of the J-domain to bind misfolded polypeptides and transfer these non-native clients to Hsp70 for refolding or degradation.Citation39,Citation40 Thus, Hsp40s select substrates for Hsp70 chaperone action and serve as the first line of defense in protein conformational disorders by recognizing non-native protein conformers. Hsp40s are classified based on the presence of several core domains found in DnaJ. Type I Hsp40s possess a J-domain, glycine/phenylaline(G/F)-rich region, and a zinc finger-like region (ZFLR) (). Type II Hsp40s possess the J-domain and G/F-rich region while Type III Hsp40s retain only the J-domain. The core Hsp40 domains described above influence Hsp40 quaternary structure and substrate selectivity ().Citation41,Citation42 Specialized Hsp40s have further acquired unique domains and modifications that likewise influence substrate preferences as well as regulation of Hsp70 refolding activity.Citation43-Citation45 For example, the yeast Type I Hsp40 Ydj1 possesses a hydrophobic depression in its C-terminal domain that binds hydrophobic peptidesCitation46 as well as a CaaX motif that is modified by farnesylation.Citation47 Interestingly, the Ydj1 ZFLR, hydrophobic polypeptide-binding pocket, and farnesyl modification all have been shown to participate in substrate binding.Citation29,Citation46,Citation48 Yet specific features are either necessary or dispensable for binding to individual substrates. Thus, Hsp40s can utilize various domain combinations to bind a wide range of non-native clients. The yeast Type II Hsp40 Sis1 also possesses a hydrophobic polypeptide-binding pocketCitation49 yet does not contain a ZFLR nor a CaaX motif. However, Sis1 does contain a G/M-rich region adjacent to the G/F-rich region,Citation50 both of which appear to influence essential cellular functions of this Hsp40.Citation51 While Ydj1 and Sis1 exhibit some overlapping physiological function,Citation52,Citation53 these Hsp40s also display distinct substrate preferencesCitation41 and as discussed below, exert very different activities on propagation of prions in yeast.
Hsp40 Activity in Propagation of [PSI+] and [URE3] Prions
Studies of the yeast prions [PSI+] and [URE3] have identified complex roles for Hsp40 co-chaperones in prion propagation and assembly into amyloid-like fibrils. The inheritable element [PSI+] is formed by the yeast translation termination factor Sup35.Citation18,Citation54 Both Ydj1 and Sis1 physically associate with large Sup35 aggregates,Citation55 though propagation of the [PSI+] prion is specifically dependent upon Sis1.Citation5,Citation30 On the other hand, overexpression of Ydj1 in conjunction with its cognate Hsp70 destabilizes “weak” [PSI+] variants.Citation6 Also noteworthy, overexpression of Apj1 (another Type I Hsp40 in yeast) cures cells of specific [PSI+] variants.Citation56 Apj1 shares strong homology with Ydj1 yet its cellular functions are still unclear. Recent studies on Sup35 fibril assembly in vitro have demonstrated a direct role for Hsp40 molecular chaperones in regulating the assembly of amyloid-like fibrils.Citation31,Citation57 Interestingly, select Hsp40:Hsp70 pairs exert different actions on Sup35 assembly as well as the prion remodeling activity of Hsp104.Citation58 Therefore, distinct chaperone complexes might selectively regulate prion assembly and propagation to alternate outcomes.
Hsp40s also regulate propagation of the yeast prion [URE3]. The [URE3] prion is formed by Ure2, a modulator of nitrogen catabolism in yeast.Citation59 Similar to [PSI+], propagation of the [URE3] prion requires Sis1Citation5 while overexpression of the Ydj1 cures yeast of [URE3].Citation16 Furthermore, Ydj1 inhibits the in vitro assembly of Ure2 into amyloid-like fibrils.Citation60,Citation61 In recent studies though, overexpression of J-domains from other yeast Hsp40s was shown to be sufficient to cure yeast of the [URE3] trait.Citation5,Citation62 These data indicate that modulating cycles of Hsp70 activity in the cell perturb [URE3] prion biogenesis and inheritance. This effect seems specific for the [URE3] prion,Citation5 suggesting this yeast prion is particularly sensitive to aberrations in prion flux through Hsp70 refolding pathways. Altogether, Sis1 and Ydj1 drive [PSI+] and [URE3] propagation to completely different outcomes whereby Sis1 promotes the efficient propagation of prion elements yet Ydj1 antagonizes this pathway. However, studies on [PSI+] and [URE3] propagation have not revealed the molecular mechanisms underlying these divergent chaperone actions.
Selective Recognition of the [RNQ+] Prion by Opposing Hsp40 Activities
Studies of the yeast prion [RNQ+] have recently revealed novel mechanisms by which Hsp40 co-chaperones bind amyloid-like prion conformers and perhaps regulate prion propagation pathways to distinct endpoints. The yeast prion [RNQ+]/[PIN+] is formed by the yeast protein Rnq1 (rich in asparagines and glutamines) ().Citation63,Citation64 The [RNQ+] state facilitates the conversion of other prions in yeastCitation64,Citation65 as well as seeding toxic conformers of an expanded glutamine form of human huntingtin.Citation66 Rnq1 possesses a C-terminal Gln/Asn-rich prion domain that is sufficient to assemble into amyloid-like fibrils in vitroCitation67,Citation68 and induce prion formation when fused in place of the Gln/Asn-rich N-terminal domain of Sup35.Citation63 The N-terminal non-prion domain of Rnq1 appears to regulate [RNQ+] prion propagation though the function of this domain is still unclear.Citation69 Not long after [RNQ+] was first described, propagation of [RNQ+] prions was shown to be dependent upon Sis1.Citation70 Deletion of other Hsp40s in yeast has no effect on the [RNQ+] state suggesting this dependency is specific for Sis1.Citation5 In contrast to Sis1, overexpression of Ydj1 cures yeast of some [RNQ+] prion variants.Citation71 Thus, similar to other yeast prions discussed above, Sis1 promotes [RNQ+] propagation while Ydj1 can inhibit [RNQ+] assembly perhaps reflecting distinct fundamental functions for these two Hsp40 co-chaperones in the prion assembly pathway ().
What might account for the opposing functions of Sis1 and Ydj1 on [RNQ+] prion propagation? Recent studies demonstrated that Sis1 and Ydj1 bind to different regions within the Rnq1 protein. For example, Sis1 binds a short, hydrophobic motif in the non-prion domainCitation12 while Ydj1 binds numerous motifs in the Gln/Asn-rich prion domain of Rnq1 ().Citation29 Interestingly, interaction between Rnq1 and either Hsp40 is dependent upon the [RNQ+] prion conformation.Citation29,Citation70 Conformational conversion of native Rnq1 into the [RNQ+] prion state might expose the Sis1-binding site in the non-prion domain. This provides a mechanism by which Sis1 action in [RNQ+] propagation is regulated through conformation-specific recognition by an Hsp40 co-chaperone. Binding between Ydj1 and the Gln/Asn-rich prion domain of Rnq1 is quite surprising because peptide array studies suggest Type I Hsp40s such as Ydj1 prefer substrates enriched in hydrophobic residues.Citation41,Citation72,Citation73 Furthermore, select binding to the Rnq1 prion domain in the [RNQ+] prion state implies that Ydj1 recognizes the Gln/Asn-rich motifs in a conformation-specific manner. Altogether, two Hsp40s in the cell bind [RNQ+] prions yet target different regions in the Rnq1 protein. The outcome of such binding preferences might (at a rudimentary level) account for the disparate chaperone activities on [RNQ+] prion propagation.
What features in Ydj1 and Sis1 direct these Hsp40 chaperones to bind distinct domains within Rnq1? Interestingly, binding between Ydj1 and the Rnq1 prion domain is dependent upon the Ydj1 ZFLR and farnesylation at its C-terminal CaaX motif.Citation29 The Ydj1 ZFLR is adjacent to two anti-parallel beta-strands that might bind the beta-rich Rnq1 prion domain through a beta-strand donor mechanism.Citation74,Citation75 How lipid modification of an Hsp40 co-chaperone contributes to substrate interaction is unclear, although farnesylation of Ydj1 has been implicated in binding to the kinase Ste11.Citation48 Thus, farnesylation appears required for binding to numerous chaperone substrates including yeast prions. In contrast, Sis1-dependent maintenance of the [RNQ+] prion state requires unique extensions in the G/F-rich region of Sis1.Citation76 These observations collectively suggest that Hsp40 co-chaperones rely on specialized modules to bind distinct domains in Rnq1 and regulate different aspects of [RNQ+] prion propagation.
Given such discrete binding preferences, how might Sis1 and Ydj1 exert their opposing activities on [RNQ+] prion propagation? Importantly, Sis1 binds to Rnq1 in a near 1:1 stoichoimetric complex while binding between Ydj1 and Rnq1 (or its Gln/Asn-rich prion domain) appears substoichiometric.Citation29,Citation76 Sis1 might coat [RNQ+] prion assemblies via binding the Rnq1 non-prion domain, direct Hsp70/Hsp104 molecular chaperones to [RNQ+] fibrils, and facilitate shearing to generate heritable [RNQ+] prion seeds.Citation28,Citation30 In addition, Sis1 might facilitate addition of new Rnq1 subunits into an elongating [RNQ+] fibril because overexpression of Sis1 increases the pool of amyloid-like [RNQ+] assemblies while overexpression of Hsp70 and Hsp104 does not result in such an increase.Citation12
In contrast to Sis1, Ydj1 might cap the exposed ends of [RNQ+] prion assemblies and thereby inhibit fibril elongation by sterically hindering contacts between exposed Gln/Asn-rich motifs in the [RNQ+] prion conformer. In addition, Ydj1 might bind a [RNQ+] prion assembly intermediate and cooperate with Hsp70 to refold the Rnq1 protein into its native conformation or partition this protein conformer into an alternative off-pathway assembly that is subsequently remodeled by another chaperone complex.Citation77 The net result of either mechanism would be solubilization of assembled Rnq1 and loss of the [RNQ+] prion trait. Importantly, Sis1 activity must normally out-compete Ydj1 to promote efficient [RNQ+] propagation. This might occur because Sis1-binding to Rnq1 is stoichiometricCitation76 and Ydj1-binding motifs in the Rnq1 prion domain are buried within most [RNQ+] assemblies. Furthermore, some but not all [RNQ+] prion variants are sensitive to Ydj1 overexpressionCitation71 suggesting that Ydj1 may differentially recognize Rnq1 prion domain surfaces exposed in specific [RNQ+] prion variants.
These studies provide novel insight on the selectivity of Hsp40 interaction with amyloid-like substrates, yet several significant questions still remain. For example, if Sis1 and Ydj1 bind distinct domains in Rnq1 do these co-chaperones bind simultaneously to the same Rnq1 protein or do Sis1 and Ydj1 act on discrete [RNQ+] intermediates/assemblies? Future studies will be required to dissect this question conclusively, yet the answer will likely reveal significant insight on the basic mechanisms of chaperone recognition of amyloid-like protein species in conformational disorders.
Hsp40s Protect Cells from Toxic Prion Conformers
The study of Hsp40 action in [RNQ+] assembly has further revealed that Sis1 and Ydj1 protect the cell from the accumulation of cytotoxic protein conformers. Overexpression of Rnq1 is toxic to yeast in the presence of pre-existing [RNQ+] prion.Citation12 Importantly, overexpression of Sis1 suppresses cytotoxicity caused by excess Rnq1, an effect that correlates with enhanced [RNQ+] prion assembly into SDS-insoluble aggregates and a decrease in the pool of SDS-soluble, Rnq1 protein species. Furthermore, mutating the Sis1-binding site in the Rnq1 non-prion domain decreases the efficiency of [RNQ+] prion assembly and exacerbates toxicity.Citation12 Thus, chaperone-mediated [RNQ+] assembly appears protective although the specific nature of the cytotoxic protein conformer is still unclear.
In contrast to full length Rnq1, overexpression of the Rnq1 prion domain alone is not toxic to yeast.Citation12 However, overexpression of this prion fragment becomes toxic in the absence of Ydj1.Citation29 Unlike full-length Rnq1, whose toxicity correlated with the appearance of a low molecular weight, soluble protein species, the toxicity by the Rnq1 prion domain in a ydj1-null background correlated with an increase in the pool of large, SDS-insoluble assemblies.Citation29 Thus, Ydj1 might be required for the cell to tolerate excessive levels of large, amyloid-like species, although this point requires further investigation. Altogether, the above observations suggest a model in which Sis1 and Ydj1 coordinate the flux of Rnq1 proteins through the [RNQ+] prion assembly pathway in order to maintain the accumulation of soluble versus amyloid-like particles within a tolerable threshold for the cell. Interestingly, Sis1 and Ydj1 were previously shown to have opposing effects on aggregation and toxicity of an expanded polyglutamine model in yeast.Citation78 Altogether, Hsp40 co-chaperones selectively bind non-native protein species to either maintain protein solubility or drive aggregation.Citation79 Even though amyloid assembly can be protective it is important to consider that excessive amyloid burden might also result in cell death.Citation80 Susceptibility to various amyloid-like protein conformers is likely dependent upon the global expression pattern of environmental factors that buffer proteotoxicity.Citation81
Concluding Remarks and Future Directions
Hsp40 molecular chaperones have recently emerged as critical regulators of prion propagation in yeast. Interestingly, individual Hsp40s modulate discrete steps in the prion assembly pathway and in the case of Ydj1 and Sis1, execute opposing activities on propagation of several yeast prions. Such disparate activities might originate from the unique binding preferences exhibited by the Hsp40 co-chaperone family, including recognition of Gln/Asn-rich regions responsible for assembly into beta-rich amyloid-like fibrils. Sis1 and Ydj1 possess unique structural domains that might account for such differential binding preferences.Citation41 For example, Ydj1 utilizes its ZFLR and a farnesyl moiety to bind the prion domain of Rnq1.Citation29 These features are conserved in various human Hsp40s and might contribute to the recognition of expanded polyglutamine conformers in various human diseases.Citation82–Citation84 Studies of Hsp40 action in [RNQ+] assembly have also demonstrated that Sis1-mediated acceleration of [RNQ+] prion assembly is cytoprotective.Citation12 Such a role for an Hsp40 co-chaperone in cytoprotection is consistent with other recent observations that facilitated protein aggregation protects against cell death mediated by Aβ(1–42)Citation85,Citation86 and the expanded polyglutamine huntingtinCitation77,Citation87,Citation88 altogether suggesting that multiple protein quality control pathways might exist to cope with the accumulation of toxic protein conformers. Thus, dissecting how Hsp40s selectively recognize toxic protein species and recruit other chaperone complexes (such as Hsp70 and Hsp104) will yield significant insight on how aggregation pathways are regulated in human conformational disorders.
Abbreviations
| Hsp | = | heat shock protein |
| Gln | = | glutamine |
| Asn | = | asparagines |
| G/F-rich | = | glycine/phenylaline-rich |
| ZFLR | = | zinc finger-like region |
| G/M-rich | = | glycine/methionine-rich |
Figures and Tables
Figure 1 Domain structures of the Type I Hsp40 Ydj1 and Type II Hsp40 Sis1 (A). Both Hsp40s possess N-terminal J-domains and adjacent G/F-rich regions. Ydj1 also contains a zinc finger-like region with two zinc-binding domains (denoted as I and II). Sis1 contains a G/M-rich region in addition to the G/F-rich region. C-terminal domains (CTD) in these Hsp40s share limited homology although both possess hydrophobic polypeptide-binding pockets (PBP) and dimerization domains (DD). Furthermore, Ydj1 is farnesylated at a C-terminal CaaX motif. (B) Quaternary structures of Ydj1 and Sis1 homodimers (for details see ref. Citation42). The polypeptide-binding pockets of each Hsp40 are noted by a small grove in the C-terminus. While the J-domains are highly conserved between Ydj1 and Sis1, the orientations of these domains with respect to the polypeptide-binding pocket are very different.
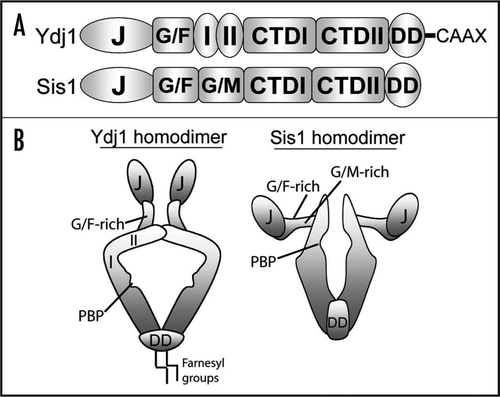
Figure 2 Domain structure of Rnq1 from S. cerevisiae (A). Rnq1 contains a N-terminal non-prion domain (aa1–152) and a C-terminal, Gln/Asn-rich prion domain (aa153–405). Sis1 binds a short, hydrophobic motif in the non-prion domain while Ydj1 binds numerous motifs in prion domain. (B) Model for Hsp40 action in [RNQ+] prion assembly pathway. Native Rnq1 is converted to the [RNQ+] prion state and assembles into large, [RNQ+] prion aggregates that are sheared to generate heritable [RNQ+] prion seeds. Sis1 might facilitate shearing to maintain a pool of [RNQ+] prion seeds thereby propagating the [RNQ+] prion state. In addition Sis1 might promote the elongation of [RNQ+] prion particles. In contrast, Ydj1 might antagonize [RNQ+] prion assembly through binding an early [RNQ+] prion conformer thus hindering [RNQ+] assembly or by accelerating the disassembly of large, [RNQ+] prions such that Rnq1 is eventually converted to its native state.
![Figure 2 Domain structure of Rnq1 from S. cerevisiae (A). Rnq1 contains a N-terminal non-prion domain (aa1–152) and a C-terminal, Gln/Asn-rich prion domain (aa153–405). Sis1 binds a short, hydrophobic motif in the non-prion domain while Ydj1 binds numerous motifs in prion domain. (B) Model for Hsp40 action in [RNQ+] prion assembly pathway. Native Rnq1 is converted to the [RNQ+] prion state and assembles into large, [RNQ+] prion aggregates that are sheared to generate heritable [RNQ+] prion seeds. Sis1 might facilitate shearing to maintain a pool of [RNQ+] prion seeds thereby propagating the [RNQ+] prion state. In addition Sis1 might promote the elongation of [RNQ+] prion particles. In contrast, Ydj1 might antagonize [RNQ+] prion assembly through binding an early [RNQ+] prion conformer thus hindering [RNQ+] assembly or by accelerating the disassembly of large, [RNQ+] prions such that Rnq1 is eventually converted to its native state.](/cms/asset/8b8e8f2c-2e47-4653-9e06-7b91374fc3f4/kprn_a_10909062_f0002.gif)
Acknowledgements
D.W.S. is supported by a pre-doctoral training grant from the National Institutes of Health (5T32GM008581-09). P.M.D. is supported by a pre-doctoral fellowship from the American Heart Association. D.M.C. is supported by funds from the National Institutes of Health (5R01GM067785-06).
References
- Tuite MF, Cox BS. Propagation of yeast prions. Nat Rev Mol Cell Biol 2003; 4:878 - 890
- Jones GW, Tuite MF. Chaperoning prions: the cellular machinery for propagating an infectious protein?. Bioessays 2005; 27:823 - 832
- Rikhvanov EG, Romanova NV, Chernoff YO. Chaperone effects on prion and nonprion aggregates. Prion 2007; 1:217 - 222
- Chernoff YO, Lindquist SL, Ono B, Inge-Vechtomov SG, Liebman SW. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+]. Science 1995; 268:880 - 884
- Higurashi T, Hines JK, Sahi C, Aron R, Craig EA. Specificity of the J-protein Sis1 in the propagation of 3 yeast prions. Proc Natl Acad Sci USA 2008; 105:16596 - 16601
- Kushnirov VV, Kryndushkin DS, Boguta M, Smirnov VN, Ter-Avanesyan MD. Chaperones that cure yeast artificial [PSI+] and their prion-specific effects. Curr Biol 2000; 10:1443 - 1446
- Schwimmer C, Masison DC. Antagonistic interactions between yeast [PSI(+)] and [URE3] prions and curing of [URE3] by Hsp70 protein chaperone Ssa1p but not by Ssa2p. Mol Cell Biol 2002; 22:3590 - 3598
- Carrell RW, Lomas DA. Conformational disease. Lancet 1997; 350:134 - 138
- Sipe JD, Cohen AS. Review: history of the amyloid fibril. J Struct Biol 2000; 130:88 - 98
- Chiti F, Dobson CM. Protein misfolding, functional amyloid and human disease. Annu Rev Biochem 2006; 75:333 - 366
- Kayed R, Head E, Thompson JL, McIntire TM, Milton SC, Cotman CW, et al. Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science 2003; 300:486 - 489
- Douglas PM, Treusch S, Ren HY, Halfmann R, Duennwald ML, Lindquist S, et al. Chaperone-dependent amyloid assembly protects cells from prion toxicity. Proc Natl Acad Sci USA 2008;
- Ross ED, Minton A, Wickner RB. Prion domains: sequences, structures and interactions. Nat Cell Biol 2005; 7:1039 - 1044
- Williams AJ, Paulson HL. Polyglutamine neurodegeneration: protein misfolding revisited. Trends Neurosci 2008; 31:521 - 528
- Orr HT, Zoghbi HY. Trinucleotide repeat disorders. Annu Rev Neurosci 2007; 30:575 - 621
- Moriyama H, Edskes HK, Wickner RB. [URE3] prion propagation in Saccharomyces cerevisiae: requirement for chaperone Hsp104 and curing by overexpressed chaperone Ydj1p. Mol Cell Biol 2000; 20:8916 - 8922
- Derkatch IL, Bradley ME, Zhou P, Chernoff YO, Liebman SW. Genetic and environmental factors affecting the de novo appearance of the [PSI+] prion in Saccharomyces cerevisiae. Genetics 1997; 147:507 - 519
- Paushkin SV, Kushnirov VV, Smirnov VN, Ter-Avanesyan MD. Propagation of the yeast prion-like [psi+] determinant is mediated by oligomerization of the SUP35-encoded polypeptide chain release factor. EMBO J 1996; 15:3127 - 3134
- Kryndushkin DS, Alexandrov IM, Ter-Avanesyan MD, Kushnirov VV. Yeast [PSI+] prion aggregates are formed by small Sup35 polymers fragmented by Hsp104. J Biol Chem 2003; 278:49636 - 49643
- Shorter J, Lindquist S. Hsp104 catalyzes formation and elimination of self-replicating Sup35 prion conformers. Science 2004; 304:1793 - 1797
- Wegrzyn RD, Bapat K, Newnam GP, Zink AD, Chernoff YO. Mechanism of prion loss after Hsp104 inactivation in yeast. Mol Cell Biol 2001; 21:4656 - 4669
- Newnam GP, Wegrzyn RD, Lindquist SL, Chernoff YO. Antagonistic interactions between yeast chaperones Hsp104 and Hsp70 in prion curing. Mol Cell Biol 1999; 19:1325 - 1333
- Jung G, Jones G, Wegrzyn RD, Masison DC. A role for cytosolic hsp70 in yeast [PSI(+)] prion propagation and [PSI(+)] as a cellular stress. Genetics 2000; 156:559 - 570
- Allen KD, Wegrzyn RD, Chernova TA, Muller S, Newnam GP, Winslett PA, et al. Hsp70 chaperones as modulators of prion life cycle: novel effects of Ssa and Ssb on the Saccharomyces cerevisiae prion [PSI+]. Genetics 2005; 169:1227 - 1242
- Chernoff YO, Newnam GP, Kumar J, Allen K, Zink AD. Evidence for a protein mutator in yeast: role of the Hsp70-related chaperone ssb in formation, stability and toxicity of the [PSI] prion. Mol Cell Biol 1999; 19:8103 - 8112
- Chacinska A, Szczesniak B, Kochneva-Pervukhova NV, Kushnirov VV, Ter-Avanesyan MD, Boguta M. Ssb1 chaperone is a [PSI+] prion-curing factor. Curr Genet 2001; 39:62 - 67
- Glover JR, Lindquist S. Hsp104, Hsp70 and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell 1998; 94:73 - 82
- Aron R, Higurashi T, Sahi C, Craig EA. J-protein co-chaperone Sis1 required for generation of [RNQ+] seeds necessary for prion propagation. EMBO J 2007; 26:3794 - 3803
- Summers DW, Douglas PM, Ren HY, Cyr DM. The Type I Hsp40 Ydj1 Utilizes a Farnesyl Moiety and Zinc Finger-like Region to Suppress Prion Toxicity. J Biol Chem 2009; 284:3628 - 3639
- Tipton KA, Verges KJ, Weissman JS. In vivo monitoring of the prion replication cycle reveals a critical role for Sis1 in delivering substrates to Hsp104. Mol Cell 2008; 32:584 - 591
- Shorter J, Lindquist S. Hsp104, Hsp70 and Hsp40 interplay regulates formation, growth and elimination of Sup35 prions. EMBO J 2008; 27:2712 - 2724
- Walsh P, Bursac D, Law YC, Cyr D, Lithgow T. The J-protein family: modulating protein assembly, disassembly and translocation. EMBO Rep 2004; 5:567 - 571
- Szabo A, Langer T, Schroder H, Flanagan J, Bukau B, Hartl FU. The ATP hydrolysis-dependent reaction cycle of the Escherichia coli Hsp70 system DnaK, DnaJ and GrpE. Proc Natl Acad Sci USA 1994; 91:10345 - 10349
- Langer T, Lu C, Echols H, Flanagan J, Hayer MK, Hartl FU. Successive action of DnaK, DnaJ and GroEL along the pathway of chaperone-mediated protein folding. Nature 1992; 356:683 - 689
- Cyr DM, Lu X, Douglas MG. Regulation of Hsp70 function by a eukaryotic DnaJ homolog. J Biol Chem 1992; 267:20927 - 20931
- Cyr DM. Swapping nucleotides, tuning Hsp70. Cell 2008; 133:945 - 947
- Sahi C, Craig EA. Network of general and specialty J protein chaperones of the yeast cytosol. Proc Natl Acad Sci USA 2007; 104:7163 - 7168
- Qiu XB, Shao YM, Miao S, Wang L. The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cell Mol Life Sci 2006; 63:2560 - 2570
- Lu Z, Cyr DM. The conserved carboxyl terminus and zinc finger-like domain of the co-chaperone Ydj1 assist Hsp70 in protein folding. J Biol Chem 1998; 273:5970 - 5978
- Fan CY, Ren HY, Lee P, Caplan AJ, Cyr DM. The type I Hsp40 zinc finger-like region is required for Hsp70 to capture non-native polypeptides from Ydj1. J Biol Chem 2005; 280:695 - 702
- Fan CY, Lee S, Ren HY, Cyr DM. Exchangeable chaperone modules contribute to specification of type I and type II Hsp40 cellular function. Mol Biol Cell 2004; 15:761 - 773
- Ramos CH, Oliveira CL, Fan CY, Torriani IL, Cyr DM. Conserved central domains control the quaternary structure of type I and type II Hsp40 molecular chaperones. J Mol Biol 2008; 383:155 - 166
- Holstein SE, Ungewickell H, Ungewickell E. Mechanism of clathrin basket dissociation: separate functions of protein domains of the DnaJ homologue auxilin. J Cell Biol 1996; 135:925 - 937
- Yan W, Schilke B, Pfund C, Walter W, Kim S, Craig EA. Zuotin, a ribosome-associated DnaJ molecular chaperone. EMBO J 1998; 17:4809 - 4817
- Lu Z, Cyr DM. Protein folding activity of Hsp70 is modified differentially by the hsp40 co-chaperones Sis1 and Ydj1. J Biol Chem 1998; 273:27824 - 27830
- Li J, Qian X, Sha B. The crystal structure of the yeast Hsp40 Ydj1 complexed with its peptide substrate. Structure 2003; 11:1475 - 1483
- Caplan AJ, Tsai J, Casey PJ, Douglas MG. Farnesylation of YDJ1p is required for function at elevated growth temperatures in Saccharomyces cerevisiae. J Biol Chem 1992; 267:18890 - 18895
- Flom GA, Lemieszek M, Fortunato EA, Johnson JL. Farnesylation of ydj1 is required for in vivo interaction with hsp90 client proteins. Mol Biol Cell 2008; 19:5249 - 5258
- Sha B, Lee S, Cyr DM. The crystal structure of the peptide-binding fragment from the yeast Hsp40 protein Sis1. Structure 2000; 8:799 - 807
- Luke MM, Sutton A, Arndt KT. Characterization of SIS1, a Saccharomyces cerevisiae homologue of bacterial dnaJ proteins. J Cell Biol 1991; 114:623 - 638
- Yan W, Craig EA. The glycine-phenylalanine-rich region determines the specificity of the yeast Hsp40 Sis1. Mol Cell Biol 1999; 19:7751 - 7758
- Caplan AJ, Douglas MG. Characterization of YDJ1: a yeast homologue of the bacterial dnaJ protein. J Cell Biol 1991; 114:609 - 621
- Johnson JL, Craig EA. An essential role for the substrate-binding region of Hsp40s in Saccharomyces cerevisiae. J Cell Biol 2001; 152:851 - 856
- Glover JR, Kowal AS, Schirmer EC, Patino MM, Liu JJ, Lindquist S. Self-seeded fibers formed by Sup35, the protein determinant of [PSI+], a heritable prion-like factor of S. cerevisiae. Cell 1997; 89:811 - 819
- Bagriantsev SN, Gracheva EO, Richmond JE, Liebman SW. Variant-specific [PSI+] Infection is Transmitted by Sup35 Polymers within [PSI+] Aggregates with Heterogeneous Protein Composition. Mol Biol Cell 2008; 19:2433 - 2443
- Kryndushkin DS, Smirnov VN, Ter-Avanesyan MD, Kushnirov VV. Increased expression of Hsp40 chaperones, transcriptional factors and ribosomal protein Rpp0 can cure yeast prions. J Biol Chem 2002; 277:23702 - 23708
- Krzewska J, Melki R. Molecular chaperones and the assembly of the prion Sup35p, an in vitro study. EMBO J 2006; 25:822 - 833
- Sweeny EA, Shorter J. Prion proteostasis: Hsp104 meets its supporting cast. Prion 2008; 2:1 - 6
- Coschigano PW, Magasanik B. The URE2 gene product of Saccharomyces cerevisiae plays an important role in the cellular response to the nitrogen source and has homology to glutathione s-transferases. Mol Cell Biol 1991; 11:822 - 832
- Lian HY, Zhang H, Zhang ZR, Loovers HM, Jones GW, Rowling PJ, et al. Hsp40 interacts directly with the native state of the yeast prion protein Ure2 and inhibits formation of amyloid-like fibrils. J Biol Chem 2007; 282:11931 - 11940
- Savistchenko J, Krzewska J, Fay N, Melki R. Molecular chaperones and the assembly of the prion URE2p in vitro. J Biol Chem 2008; 283:15732 - 15739
- Sharma D, Stanley RF, Masison DC. Curing of Yeast [URE3] Prion by the Hsp40 Cochaperone Ydj1p Is Mediated by Hsp70. Genetics 2009; 181:129 - 137
- Sondheimer N, Lindquist S. Rnq1: an epigenetic modifier of protein function in yeast. Mol Cell 2000; 5:163 - 172
- Derkatch IL, Bradley ME, Hong JY, Liebman SW. Prions affect the appearance of other prions: the story of [PIN(+)]. Cell 2001; 106:171 - 182
- Derkatch IL, Uptain SM, Outeiro TF, Krishnan R, Lindquist SL, Liebman SW. Effects of Q/N-rich, polyQ and non-polyQ amyloids on the de novo formation of the [PSI+] prion in yeast and aggregation of Sup35 in vitro. Proc Natl Acad Sci USA 2004; 101:12934 - 12939
- Meriin AB, Zhang X, He X, Newnam GP, Chernoff YO, Sherman MY. Huntington toxicity in yeast model depends on polyglutamine aggregation mediated by a prion-like protein Rnq1. J Cell Biol 2002; 157:997 - 1004
- Vitrenko YA, Gracheva EO, Richmond JE, Liebman SW. Visualization of aggregation of the Rnq1 prion domain and cross-seeding interactions with Sup35NM. J Biol Chem 2007; 282:1779 - 1787
- Wickner RB, Dyda F, Tycko R. Amyloid of Rnq1p, the basis of the [PIN+] prion, has a parallel in-register beta-sheet structure. Proc Natl Acad Sci USA 2008; 105:2403 - 2408
- Kurahashi H, Ishiwata M, Shibata S, Nakamura Y. A regulatory role of the Rnq1 nonprion domain for prion propagation and polyglutamine aggregates. Mol Cell Biol 2008; 28:3313 - 3323
- Sondheimer N, Lopez N, Craig EA, Lindquist S. The role of Sis1 in the maintenance of the [RNQ+] prion. EMBO J 2001; 20:2435 - 2442
- Bradley ME, Edskes HK, Hong JY, Wickner RB, Liebman SW. Interactions among prions and prion “strains” in yeast. Proc Natl Acad Sci USA 2002; 99:16392 - 16399
- Li J, Sha B. Peptide substrate identification for yeast Hsp40 Ydj1 by screening the phage display library. Biol Proced Online 2004; 6:204 - 208
- Rudiger S, Schneider-Mergener J, Bukau B. Its substrate specificity characterizes the DnaJ co-chaperone as a scanning factor for the DnaK chaperone. EMBO J 2001; 20:1042 - 1050
- Sauer FG, Futterer K, Pinkner JS, Dodson KW, Hultgren SJ, Waksman G. Structural basis of chaperone function and pilus biogenesis. Science 1999; 285:1058 - 1061
- Choudhury D, Thompson A, Stojanoff V, Langermann S, Pinkner J, Hultgren SJ, et al. X-ray structure of the FimC-FimH chaperone-adhesin complex from uropathogenic Escherichia coli. Science 1999; 285:1061 - 1066
- Lopez N, Aron R, Craig EA. Specificity of class II Hsp40 Sis1 in maintenance of yeast prion [RNQ+]. Mol Biol Cell 2003; 14:1172 - 1181
- Behrends C, Langer CA, Boteva R, Bottcher UM, Stemp MJ, Schaffar G, et al. Chaperonin TRiC promotes the assembly of polyQ expansion proteins into nontoxic oligomers. Mol Cell 2006; 23:887 - 897
- Gokhale KC, Newnam GP, Sherman MY, Chernoff YO. Modulation of prion-dependent polyglutamine aggregation and toxicity by chaperone proteins in the yeast model. J Biol Chem 2005; 280:22809 - 22818
- Douglas PM, Summers DW, Cyr DM. Molecular chaperones antagonize proteotoxicity by differentially modulating protein aggregation pathways. Prion 2009; 3
- Fiala JC. Mechanisms of amyloid plaque pathogenesis. Acta Neuropathol 2007; 114:551 - 571
- Balch WE, Morimoto RI, Dillin A, Kelly JW. Adapting proteostasis for disease intervention. Science 2008; 319:916 - 919
- Chai Y, Koppenhafer SL, Bonini NM, Paulson HL. Analysis of the role of heat shock protein (Hsp) molecular chaperones in polyglutamine disease. J Neurosci 1999; 19:10338 - 10347
- Chan HY, Warrick JM, Gray-Board GL, Paulson HL, Bonini NM. Mechanisms of chaperone suppression of polyglutamine disease: selectivity, synergy and modulation of protein solubility in Drosophila. Hum Mol Genet 2000; 9:2811 - 2820
- Cummings CJ, Mancini MA, Antalffy B, DeFranco DB, Orr HT, Zoghbi HY. Chaperone suppression of aggregation and altered subcellular proteasome localization imply protein misfolding in SCA1. Nat Genet 1998; 19:148 - 154
- Cohen E, Bieschke J, Perciavalle RM, Kelly JW, Dillin A. Opposing activities protect against age-onset proteotoxicity. Science 2006; 313:1604 - 1610
- Cheng IH, Scearce-Levie K, Legleiter J, Palop JJ, Gerstein H, Bien-Ly N, et al. Accelerating amyloid-beta fibrillization reduces oligomer levels and functional deficits in Alzheimer disease mouse models. J Biol Chem 2007; 282:23818 - 23828
- Wyttenbach A, Carmichael J, Swartz J, Furlong RA, Narain Y, Rankin J, et al. Effects of heat shock, heat shock protein 40 (HDJ-2), and proteasome inhibition on protein aggregation in cellular models of Huntington's disease. Proc Natl Acad Sci USA 2000; 97:2898 - 2903
- Wacker JL, Zareie MH, Fong H, Sarikaya M, Muchowski PJ. Hsp70 and Hsp40 attenuate formation of spherical and annular polyglutamine oligomers by partitioning monomer. Nat Struct Mol Biol 2004; 11:1215 - 1222