Abstract
Prion diseases, which are called transmissible spongiform encephalopathies (TSE), comprise a group of fatal infectious neurodegenerative disorders. Investigation of prion strains and generation of species dependent TSE model are necessary to understand pathogenesis of the disease. To establish a BSE-specific in vitro cell culture model, N2a and GT1 mouse neuronal cell lines were generated to express the bovine prion protein by transfection of the bovine prion gene (Prnp). In addition, the endogenous mouse prion protein was suppressed in N2a, NbP, GT1, and GbP cell lines using the siRNA duplexes, siRNA1 and siRNA2 that target the N- and C- termini of murine Prnp, respectively. Both siRNA1 and siRNA2 effectively decreased murine prion protein levels by more than 80% and the down-regulation efficacy was increased in siRNA dose-dependent manner. The greatest down-regulation was observed 48 h after siRNA delivery. The moPrnp knockdown NbP and GbP cell lines and the Prnp-targeting siRNA technique established in the present study would be useful tools for dissecting the basic mechanisms of prion infection, especially for BSE.
Introduction
Transmissible spongiform encephalopathies (TSEs), or prion diseases, are inevitably fatal, and there is no effective therapy.1 Although prion diseases have been recognized for decades, or even centuries, important questions still remain regarding the nature of the infectious agents, the mechanisms of prion replication, the molecular properties underlying prion strains, and the mechanistic basis of species barriers.Citation1,Citation2 Two properties of TSEs are particularly perplexing. The first is the existence of multiple prion strains that produce distinct disease phenotypes within the same animal species.Citation3,Citation4 The second is the species barrier that restricts transmission of prion diseases between different mammalian species.Citation5 There are several strains that cause TSE. Each strain has distinguishing characteristics, in particular by their incubation periods and neuropathological profiles when passaged into panels of inbred mice.Citation4,Citation6,Citation7 Most prion researchers have investigated TSE in mouse and hamster experimental models.
RNA interference (RNAi) provides a rapid way of depleting mRNA through the introduction of double-stranded RNA that is homologous to a particular message, leading to the degradation of the targeted message.Citation8–Citation10 This technique has been adopted by the research community as a versatile tool with a wide range of applications.Citation11,Citation12 The utility of RNAi has resulted in an explosion of interest in deciphering the molecular mechanismsCitation9,Citation13 behind such processes as oncogene repression in cancer,Citation14 cell signalingCitation15 and the development of novel therapeutics.Citation16,Citation17 In the field of neurodegenerative disorders, several RNAi-based studies have focused on developing therapies that knockdown Prnp of a disease-causing mutated gene.Citation8,Citation13,Citation16,Citation18,Citation19
One solution considering various species and strains in prion biology is an establishment of new research models. To generate a BSE-specific in vitro cell culture model, N2a and GT1 mouse neuronal cells were generated to express the exogenous bovine prion protein (boPrP) through transfection of the bovine prion gene (boPrnp). Then, small interfering RNA (siRNA) targeting the endogenous mouse prion gene (moPrnp) were designed, and the siRNA-mediated knockdown of mouse prion protein (moPrP) was evaluated in each of the cell lines.
Results
Generation of stable cell lines, NbP and GbP, expressing the bovine prion protein.
The insertion of boPrnp was confirmed through genomic PCR and compared to the cells transfected with the empty vector and to wild-type cells. By genomic PCR, NbP and GbP tested positive for the 1,119 bp amplicon containing the multiple cloning site (MCS) of the vector plus the inserted boPrnp, while cells transfected with the empty vector yielded only the 300 bp amplicon containing the MCS of the empty vector. No amplification occurred with genomic DNA from wild-type N2a and GT1 cells (data not shown).
Expression of endogenous murine Prnp gene was dose-dependently downregulated while the expression of bovine Prnp gene was constant regardless siRNA concentration (). RT-PCR was performed to detect the transcripts of the derived gene using boPrnp-specific primer pairs. The 819 bp amplicons with similar density were observed in N2a and GT1 cells harboring the boPrnp ORF (NbP and GbP), while there were no amplicons from cells transfected with empty vector or from wild-type cells. No DNA contamination was observed in the RNA samples, as verified by repeating the above procedure with reverse transcriptase omitted (data not shown).
Efficient downregulation of endogenous moPrP expression using siRNAs.
The knockdown efficiency of siRNA1, which targets the N-terminus of moPrnp, was investigated through one-step real time RT-PCR. A standard curve was calculated using linear regression analysis. This standard curve displayed a linear relationship between threshold cycle (Ct) values and the log copy number of the input molecules: Ct = −2.5114 Log (CopyNr) + 36.956 (R2 = 0.9717). The corresponding copy number of the PCR amplified moPrnp ORF was calculated using the equation 1 μg of 1,000 bp DNA = 9.1 × 1011 molecules.Citation20 The amount of product in a particular sample was determined from the standard curve of Ct values. The target mouse prion mRNA was dramatically reduced in dose-dependent manner by the introduction of siRNA1 into NbP and GbP cells. The observed percentage of inhibition ranged from 88.6 to 98.5% in NbP cells and from 74.0 to 99.6% in GbP cells following transfection of 20 to 400 nM of siRNA1, respectively ().
The downregulation of endogenous moPrP and the stable expression of exogenous boPrP were verified by western blot analysis using mouse PrP specific-3F10 and both PrP detecting-SAF70 antibodies. In the analysis with moPrP specific-3F10, expression level of moPrP was 30.9 and 50.3% in NbPsiRNA and GbPsiRNA cells respectively compared with moPrP of wild type cells. However, with SAF70 in these cells, total PrP including moPrP and boPrP was not downregulated and showed 97.9 and 127.4% of prion protein because boPrP was stably expressed regardless of anti moPrnp-siRNA treatment. Likewise, total prion protein in NbP or GbP cells showed the highest level because moPrP and boPrP displayed simultaneously in these cells ().
Decreased expression of prion protein was observed in each cell line treated with siRNA1 or siRNA2 targeting the N- or C-terminus of the mouse prion gene, respectively, compared with the non-treated control. Densitometric evaluation of the western blot demonstrated a 66.6 to 98.7% downregulation from the transfection of 200 nM siRNA1 and a 38.6 to 92.4% downregulation from 100 nM siRNA1. In the case of siRNA2, the knockdown ranged from 92.6 to 99.2% at a concentration of 200 nM and from 41.0 to 60.6% at a concentration of 100 nM (). The highest downregulation was observed at 48 h after transfection with a 94.9% downregulation at 100 nM siRNA1 and a 96.6% downregulation at 100 nM siRNA2 in N2a cells ().
Discussion
To establish BSE-specific cell culture models, mouse neuronal cells were transfected with boPrnp and then moPrP was downregulated by moPrnp specific siRNA. N2a and GT1 cells were selected because only a few cell lines, including these cell lines and the rat adrenal phaeochromocytoma line PC12, can propagate scrapie prions over many passages.Citation21 The successful downregulation of prion protein in several species, including mice, has been reported previously.Citation16,Citation18,Citation19,Citation22 However, the siRNA target sequences for moPrnp designed previously could not be applied to this experiment because they recognized or very closely matched with boPrnp sequences.Citation19 It is obviously important to verify that an siRNA which is fully homologous to the desired mRNA target has as little homology as possible to other mRNAs.Citation10 Therefore, the coding sequence of the moPrnp ORF was processed with a computer algorithm and the siRNA output was screened by alignment with boPrnp ORF to avoid redundant sequences.
Successful RNAi experiments depend on the effect of delivery system for siRNA into cells. Variable and low transfection efficiency can be one of limiting factors in gene silencing experiments, particularly with difficult-to-transfect cells such as primary neurons or suspension cells.Citation10,Citation23 The siRNA duplexes labeled with fluorescent dyes are frequently used to monitor transfection efficiency, and can allow the optimization of experimental conditions. The siRNA transfection efficiency in this study was approximately 2 times greater than a previous report showing an 82% knockdown at a fluorescenin siRNA concentration of 400 nM in N2a cells.Citation22
In N2a and GT1 cells, both siRNA1 and siRNA2 effectively decreased prion protein expression by more than 80%. This downregulation was associated with a concomitant reduction of the mRNA level by real-time RT-PCR demonstrating that, as expected, the siRNA-mediated regulation was mainly due to the degradation of target mRNA rather than inhibition of its translation. The siRNA-mediated knockdown peaked at 48 h after the transfection and gradually decreased over time as a consequence of cell division and/or degradation of the siRNA molecules.24 However, similar density of boprnp amplicon in RT-PCR and western blot analysis with mouse specific or total prion detecting antibodies suggested constant expression of exogenous boPrnp transcripts as well as boPrP regardless of anti moPrnp-siRNA treatment. In the exogenous delivery of siRNA, the significant concern is the cytotoxic side-effects to target cells.Citation25 Cytotoxic effects were increased at siRNA concentrations above 200 nM, and knockdown efficiency was positively related with the siRNA concentration in this experiment.
The newly generated NbP and GbP cell lines in this study and the applied siRNA technique would be useful for elucidating the basic mechanisms of prion pathogenesis, especially in BSE. Even if knockout strategies have been applied to reduce the gene expression for different purposes, these intergenic splicing events caused ectopic gene expression of the doppel (Dpl) protein encoded by the Prnd gene, placing the Dpl locus under the control of the Prnp promoter.Citation26,Citation27 Although untested, Dpl expression is probably not under the control of the siRNA machinery because siRNA is a post-transcriptional gene silencing mechanism. Reducing PrPC expression is one of potential therapeutic approaches to delaying the onset of prion diseases.Citation1,Citation8 Together with liposomally formulated siRNA molecules, siRNA-lipoplexes,Citation28 or magnetic nanoparticles labeled with a near-infrared dye and covalently linked to siRNA molecules,Citation12,Citation29 the cell lines and siRNA technique presented in this study could potentially be used as therapeutic agents for prion diseases. Further vector-mediated siRNA studies are needed to determine siRNA targeting sites and appropriate delivery systems.
Materials and Methods
N2a and GT1 cell cultures.
N2a, a mouse neuroblastoma cell line, and GT1, a mouse hypothalamic cell line, were maintained in Dulbecco’s modified Eagle’s Medium (DMEM) with high glucose (4.5 g/L) and 2 mM glutamine (Gibco, USA) supplemented with 10% fetal bovine serum, penicillin and streptomycin. Cells were cultured at 37°C in a 5% CO2 incubator.
Construction of the boPrnp expression vector.
The Prnp ORF of Bos taurus coreanae (a breed of cattle native to Korea) encodes a protein of 264 amino acids with a predicted molecular weight of 28 kD.Citation30 This ORF was cloned into the pIRESpuro3 eukaryotic expression vector (Clontech, USA). Primers were designed to insert the EcoRI restriction site into the flanking regions of boPrnp (AF517842) (). To dephosphorylate the linearized vector, 1 μL of calf intestinal phosphatase (CIP, Roche, Germany) and 10x CIP buffer were added and incubated at 37°C for 1 h. The gel-purified PCR product and the dephosphorylated, linearized vector were ligated with T4 ligase and transformed into DH5α competent cells (RBC, Taiwan). Successful subcloning of boPrnp was confirmed by DNA sequencing (ABI PRISM 377x L, Perkin Elmer, USA) after colony PCR and EcoRI digestion.
Transfection and generation of stable cell lines, NbP and GbP, expressing the bovine prion protein.
To generate cells stably expressing boPrP, N2a and GT1 cells (referred to NbP and GbP, respectively) were transfected with 4 μg pIRESpuro3-boPrnp using the FuGENE6 transfection reagent (Roche, Germany). Clonal lines were developed by limiting dilution and subsequent puromycin selection.
Selection of target sequences for siRNA.
siRNA was designed to knockdown the endogenous Mus musculus prion protein (moPrP) (accession number NM_011170) in each cell line. An siRNA design algorithm provided by the Invitrogen web-site (USA)Citation31 was used to select target sequences. The basic local alignment search tool (BLAST) was used against the mouse genome database to identify unique regions in the murine Prnp (moPrnp) ORF. Two regions were chosen and two siRNAs that targeted the N- and C-termini of the moPrnp ORF, had a 52% GC content, and did not recognize the boPrnp ORF were designed ().
Transfection of siRNA.
Early passage cells were transfected with siRNA duplexes at a final concentration of 200, 100 and 0 nM. After a 48 h incubation, the knockdown efficiency was assessed by real time RT-PCR and western blot. To assess the time-dependent inhibition of moPrP, N2a cells were transfected with 100 nM of siRNA, and moPrP knockdown was determined by western blot.
Genomic PCR.
Primer pairs were used at a final concentration of 0.5 pmol/μL and were designed to amplify the 819 bp boPrnp. Another primer pair which targets a 300 bp region, including the MCS of the vector, was used to screen for gene insertion into cells (Table 1). The 20 μL PCR reactions included 0.3 μg of genomic DNA, 0.5 μL of each primer, 0.2 μL of Taq DNA polymerase (5 U/μL), 2 μL of 10x PCR buffer, 2 μL of dNTP mixture (2.5 mM of each), and 14.8 μL of autoclaved distilled water. The PCR parameters consisted of an initial denaturation step at 95°C for 5 min followed by 40 cycles of denaturation at 95°C for 30 sec, annealing at 60°C for 30 sec, and extension at 72°C for 1 min and a final extension at 72°C for 15 min.
RT-PCR.
Single-stranded cDNA was synthesized by one-step RT-PCR with a reverse transcriptase and Taq DNA polymerase mixture (Invitrogen, USA). Fifty μL of the RT-PCR reaction included 25 μL of 2x reaction mix (0.4 mM of each dNTP, 2.4 mM MgSO4), 5 μg of isolated total RNA, 1 μL of each boPrnp-specific primer at a final concentration of 0.4 pmol/uL (), 1 μL of the reverse transcriptase and Taq DNA polymerase mixture, and 22 μL of autoclaved distilled water. To test for DNA contamination of the RNA samples, control reactions were prepared by omitting the reverse transcriptase. Thermal cycling for the RT-PCR was performed as follows: 30 min at 50°C for reverse transcription, an initial 2 min denaturation at 94°C; 40 cycles of denaturation at 94°C for 30 sec, annealing at 60°C for 30 sec, and extension at 72°C for 1 min; and a final extension at 72°C for 15 min.
Quantitative real-time RT-PCR.
The moPrnp ORF-specific primers and probes for real-time RT-PCR were designed using Primer Express software (Applied Biosystems, UK) (Table 1) and did not recognize boPrnp. The TaqMan minor groove binder (MGB) probe (Applied Biosystems, UK) was labeled with a 5′ reporter dye, 6-carboxyfluorescein (FAM) and a 3′ nonfluorescent quencher (NFQ) plus MGB. The 25 μL RT-PCR reaction included 11 μL of RNase-free water, 5 μL of 5x TaqMan EZ buffer (250 mM Bicine, 575 mM potassium acetate, 0.05 mM EDTA, 300 nM Passive ref. 1, 40% glycerol, pH 8.2), 3 μL of manganese acetate (25 mM Mn(OAc)2), 2.8 μL of dNTP/dUTP mix (10 mM each of dATP, dCTP, dGTP and 20 mM of dUTP), 2 μL of probe mix (18 μM of each primer and 5 μM probe), 1 μL of DNA polymerase (2.5 U/μL of DNA polymerase in 20 mM Tris-HCl, pH 8.0, 100 mM KCl, 0.1 mM EDTA, 1 mM DTT, 50% glycerol, 0.5% Tween 20), and 0.2 μL of AmpErase UNG (1 U/μL uracil N-glycosylase in 30 mM Tris-HCI, pH 7.5, 1 mM EDTA, 150 mM NaCl, 1 mM DTT, 5% glycerol, 0.5% Tween 20). RNA input was normalized with the TaqMan mouse GAPDH endogenous control (Applied Biosystems, UK). Thermal cycling for the TaqMan 7000 was performed at 2 min at 50°C for reverse transcription, 10 min at 95°C for heat inactivation of the reverse transcriptase and activation of the DNA polymerase, 50 cycles of 15 sec at 95°C, and 1 min at 60°C. The fluorescence data were collected at the end of the 60°C step using the 7000 system SDS v1.2.3 (Applied Biosystems, UK). Ten-fold serial dilutions of the PCR amplification products with the moPrnp ORF-specific primer set were used to generate a standard curve ().
Western blot analysis.
For western blot analysis, 5 μg protein from each cell lysate was prepared and separated on a 12% Tris-glycine gel and transferred to a 0.2 micron nitrocellulose membrane using a dry blotting system (Invitrogen, USA). The membrane was blocked with 5% nonfat dry milk in TritonX-tris buffered saline (TTBS: 0.1% TritonX-100 in 100 mM Tris-HCl, pH 7.5, 0.9% NaCl) for 2 h at room temperature and then probed with 3F10 which recognises only mouse PrP at a dilution of 1:10,000;Citation32 and SAF70 (Sapphire Bioscience, Australia) which cross reacts with mouse and bovine PrP at a dilution of 1:5,000. Membranes were incubated overnight at 4°C with the primary antibody. Anti-mouse IgG secondary-conjugated horseradish peroxidase (Santa Cruz, USA) diluted at 1:5,000 was then added. The bound antibodies were visualized by chemiluminescence (Amersham, UK) and protein levels were evaluated through densitometry (Multi Gauge v2.3 software, Fujifilm, Japan). The membrane was washed twice with TTBS and put in stripping solution (100 mM Tris-HCl, pH 7.5, 0.9% NaCl, 7 μL/mL β-mercaptoethanol, 2% SDS) at 50°C for 15 min. The stripped membrane was blocked with 5% nonfat dry milk in TTBS and probed with anti-mouse GAPDH (Santa Cruz, USA) at a dilution of 1:5,000 as a cellular marker. After an overnight incubation at 4°C, anti-mouse IgG-conjugated horseradish peroxidase diluted at 1:5,000 was added. Protein levels were visualized as described above.
Abbreviations
| TSE | = | transmissible spongiform encephalopathy |
| BSE | = | bovine spongiform encephalopathy |
| GT1 | = | mouse hypothalamic cell line |
| GbP | = | GT1 harboring bovine Prnp gene |
| N2a | = | mouse neuroblastoma cell line |
| NbP | = | N2a harboring bovine Prnp |
| moPrnp/boPrnp | = | mouse/bovine prion gene |
| moPrP/boPrP | = | mouse/bovine prion protein |
| RNAi | = | RNA interference |
| siRNA | = | small interfering RNA |
Figures and Tables
Figure 1 Rreal-time RT-PCR analysis to determine the level of mouse prion mRNA (A) and RT-PCR analysis to determine the expression of bovine prion mRNA (B) in NbP and GbP cells after transfection with different concentrations of siRNA. The quantity of mouse prion mRNA was normalized to that of GAPDH. An increase in siRNA resulted in a gradual suppression of the mouse prion mRNA, while the expression of bovine prion mRNA remained unchanged.
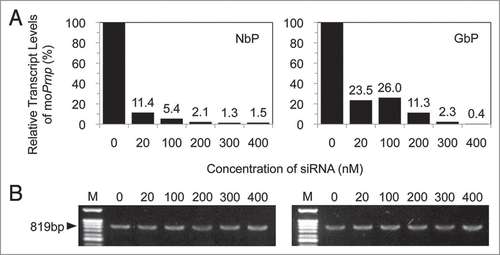
Figure 2 Western blot analysis to evaluate the abundance of mouse prion protein with 3F10 and prion proteins including moPrP and boPrP with SAF70 in each cell line after transfection of 100 nM of siRNA. Densitometric evaluation showed downregulated moPrP and stable expressed boPrP in NbPsiRNA cells (A) or GbPsiRNA cells (B) compared with siRNA non-treated cells.
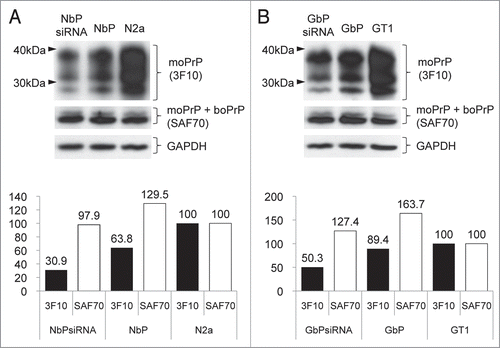
Figure 3 Downregulation of prion expression by different concentrations of siRNA1 or siRNA2. Cells were transfected with siRNA1 (A) or siRNA2 (B) and incubated for 48 h. The level of the expressed prion protein was determined by western blot (above) and evaluated by densitometry (below). Increasing concentrations of siRNA resulted in a gradual suppression of prion protein expression.
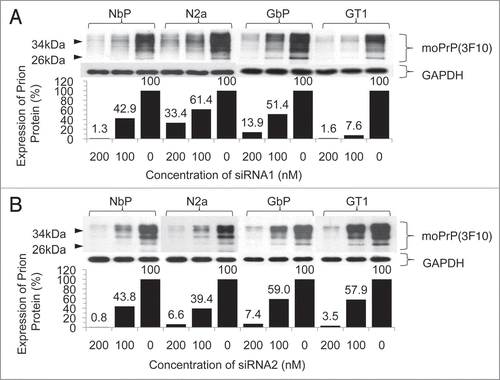
Figure 4 Time-dependent inhibition of prion expression by siRNA-mediated knockdown. Cells were transfected with 100 nM siRNA. The levels of prion expression were determined by western blot (above) and evaluated by densitometry (below). The downregulation by each siRNA was effective within 72 h of transfection by more than 80%.
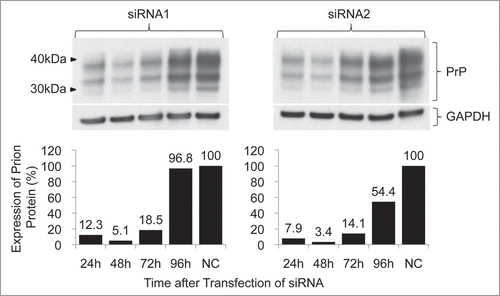
Table 1 Oligonucleotide sequences of primers, probe and siRNAs used in this study
References
- Aguzzi A, Heikenwalder M. Pathogenesis of prion diseases: current status and future outlook. Nature Rev 2006; 4:765 - 775
- Chien P, Weissman JS, DePace AH. Emerging principles of conformation-based prion inheritance. Ann Rev Biochem 2004; 73:617 - 656
- Beringue V, Bencsik A, Le Dur A, Reine F, Lai TL, Chenais N, et al. Isolation from cattle of a prion strain distinct from that causing bovine spongiform encephalopathy. PLoS Pathog 2006; 2:112
- Capobianco R, Casalone C, Suardi S, Mangieri M, Miccolo C, Limido L, et al. Conversion of the BASE Prion Strain into the BSE Strain. The Origin of BSE PLoS Pathog 2007; 3:31
- Vanik DL, Surewicz KA, Surewicz WK. Molecular basis of barriers for interspecies transmissibility of mammalian prions. Mol Cell 2004; 14:139 - 145
- Rigter A, Langeveld JP, Timmers-Parohi D, Jacobs JG, Moonen PL, Bossers A. Mapping of possible prion protein self-interaction domains using peptide arrays. BMC Biochem 2007; 8:6
- Collinge J, Sidle KC, Meads J, Ironside J, Hill AF. Molecular analysis of prion strain variation and the aetiology of ‘new variant’ CJD. Nature 1996; 383:685 - 690
- Kong Q. RNAi: a novel strategy for the treatment of prion diseases. J Clin Invest 2006; 116:3101 - 3103
- Sen GL, Blau HM. A brief history of RNAi: the silence of the genes. Faseb J 2006; 20:1293 - 1299
- Cullen BR. Enhancing and confirming the specificity of RNAi experiments. Nat Methods 2006; 3:677 - 681
- Aigner A. Delivery Systems for the Direct Application of siRNAs to Induce RNA Interference (RNAi) In Vivo. J Biomed Biotechnol 2006; 2006:71659
- Li CX, Parker A, Menocal E, Xiang S, Borodyansky L, Fruehauf JH. Delivery of RNA interference. Cell Cycle 2006; 5:2103 - 2109
- Ralph GS, Radcliffe PA, Day DM, Carthy JM, Leroux MA, Lee DC, et al. Silencing mutant SOD1 using RNAi protects against neurodegeneration and extends survival in an ALS model. Nat Medicine 2005; 11:429 - 433
- Butz K, Ristriani T, Hengstermann A, Denk C, Scheffner M, Hoppe-Seyler F. siRNA targeting of the viral E6 oncogene efficiently kills human papillomavirus-positive cancer cells. Oncogene 2003; 22:5938 - 5945
- Zeng P, Wagoner HA, Pescovitz OH, Steinmetz R. RNA interference (RNAi) for extracellular signal-regulated kinase 1 (ERK1) alone is sufficient to suppress cell viability in ovarian cancer cells. Cancer Biol Ther 2005; 4:961 - 967
- Pfeifer A, Eigenbrod S, Al-Khadra S, Hofmann A, Mitteregger G, Moser M, et al. Lentivector-mediated RNAi efficiently suppresses prion protein and prolongs survival of scrapie-infected mice. J Clin Invest 2006; 116:3204 - 3210
- Wall NR, Shi Y. Small RNA: can RNA interference be exploited for therapy. Lancet 2003; 362:1401 - 1403
- Golding MC, Long CR, Carmell MA, Hannon GJ, Westhusin ME. Suppression of prion protein in livestock by RNA interference. Pro Nat Acad Sci 2006; 103:5285 - 5290
- Tilly G, Chapuis J, Vilette D, Laude H, Vilotte JL. Efficient and specific downregulation of prion protein expression by RNAi. Biochem Biophys Res Commun 2003; 305:548 - 551
- Overbergh L, Giulietti A, Valckx D, Decallonne R, Bouillon R, Mathieu C. The use of real-time reverse transcriptase PCR for the quantification of cytokine gene expression. J Biomol Tech 2003; 14:33 - 43
- Weissmann C. The state of the prion. Nat Rev Microbiol 2004; 2:861 - 871
- Daude N, Marella M, Chabry J. Specific inhibition of pathological prion protein accumulation by small interfering RNAs. J Cell Sci 2003; 116:2775 - 2779
- Krichevsky AM, Kosik KS. RNAi functions in cultured mammalian neurons. Pro Nat Acad Sci 2002; 99:11926 - 11929
- Clark J, Ding S. Generation of RNAi Libraries for High-Throughput Screens. J Biomed Biotechnol 2006; 2006:45716
- Sioud M. Therapeutic siRNAs. Trends Pharmacol Sci 2004; 25:22 - 28
- Qin K, Zhao L, Tang Y, Bhatta S, Simard JM, Zhao RY. Doppel-induced apoptosis and counteraction by cellular prion protein in neuroblastoma and astrocytes. Neuroscience 2006; 141:1375 - 1388
- Sakudo A, Lee DC, Nakamura I, Taniuchi Y, Saeki K, Matsumoto Y, et al. Cell-autonomous PrP-Doppel interaction regulates apoptosis in PrP gene-deficient neuronal cells. Biochem Biophys Res Commun 2005; 333:448 - 454
- Santel A, Aleku M, Keil O, Endruschat J, Esche V, Durieux B, et al. RNA interference in the mouse vascular endothelium by systemic administration of siRNA-lipoplexes for cancer therapy. Gene Therapy 2006; 13:1360 - 1370
- Medarova Z, Pham W, Farrar C, Petkova V, Moore A. In vivo imaging of siRNA delivery and silencing in tumors. Nat Medicine 2007; 13:372 - 377
- Kang SG, Kang SK, Lee DY, Park YH, Hwang WS, Yoo HS. Cloning, Sequencing and Expression of cDNA Encoding Bovine Prion Protein. J Microbiol Biotechnol 2004; 14:417 - 421
- Smith C. Sharpening the tools of RNA interference. Nat Methods 2006; 3:475 - 486
- Choi JK, Park SJ, Jun YC, Oh JM, Jeong BH, Lee HP, et al. Generation of monoclonal antibody recognized by the GXXXG motif (glycine zipper) of prion protein. Hybridoma 2006; 25:271 - 277