Abstract
The prion hypothesis1-3 states that the prion and non-prion form of a protein differ only in their 3D conformation and that different strains of a prion differ by their 3D structure.4, 5 Recent technical developments have enabled solid-state NMR to address the atomic-resolution structures of full-length prions, and a first comparative study of two of them, HET-s and Ure2p, in fibrillar form, has recently appeared as a pair of companion papers.6, 7 Interestingly, the two structures are rather different: HET-s features an exceedingly well-ordered prion domain and a partially disordered globular domain. Ure2p in contrast features a very well ordered globular domain with a conserved fold, and – most probably - a partially ordered prion domain.6 For HET-s, the structure of the prion domain is characterized at atomic-resolution. For Ure2p, structure determination is under way, but the highly resolved spectra clearly show that information at atomic resolution should be achievable.
Despite the large interest in the basic mechanisms of fibril formation and prion propagation, little is known about the molecular structure of prions at atomic resolution and the mechanism of propagation. Prions with related properties to the ones responsible for mammalian diseases were also discovered in yeast and funghiCitation8,Citation9 which provide convenient model system for their studies. Prion proteins described include the mammalian prion protein PrP, Ure2p,Citation10 Rnq1p,Citation11 Sup35,Citation12 Swi1,Citation13 and Cyc8,Citation14 from bakers yeast (S. cervisiae) and HET-s from the filamentous fungus P. anserina. The soluble non-prion form of the proteins characterized in vitro is a globular protein with an unfolded, dynamically disordered N- or C-terminal tail.Citation15–Citation18 In the prion form, the proteins form fibrillar aggregates, in which the tail adopts a different conformation and is thought to be the dominant structural element for fibril formation.
Fibrills are difficult to structurally characterize at atomic resolution, as X-ray diffraction and liquid-state NMR cannot be applied because of the non-crystallinity and the mass of the fibrils. Solid-state NMR, in contrast, is nowadays well suited for this purpose. The size of the monomer, between 230 and 685 amino-acid residues for the prions of , and therefore the number of resonances in the spectrum—that used to be large for structure determination—is now becoming tractable by this method.
Prion proteins characterized so far were found to be usually constituted of two domains, namely the prion domain and the globular domain (see ). This architecture suggests a divide-and-conquer approach to structure determination, in which the globular and prion domain are investigated separately. In isolation, the latter, or fragments thereof, were found to form β-sheet rich structures (e.g., Ure2p(1-89),Citation6,Citation19 Rnq1p(153-405)Citation20 and HET-s(218-289)Citation21). The same conclusion was reached by investigating Sup35(1-254).Citation22 All these fragements have been characterized as amyloids, which we define in the sense that a significant part of the protein is involved in a cross-beta motif.Citation23 An atomic resolution structure however is available presently only for the HET-s prion domain, and was obtained from solid-state NMRCitation24 (vide infra). It contains mainly β-sheets, which form a triangular hydrophobic core. While this cross-beta structure can be classified as an amyloid, its triangular shape does deviate significantly from amyloid-like structures of smaller peptides.Citation23
Regarding the globular domains, structures have been determined by x-ray crystallography (Ure2pCitation25,Citation26 and HET-sCitation27), as well as NMR (mammal prionsCitation15,Citation28–Citation30). All reveal a protein fold rich in α-helices, and dimeric structures for the Ure2 and HET-s proteins. The Ure2p fold resembles that of the β-class glutathione S-transferases (GST), but lacks GST activity.Citation25
It is a central question for the structural biology of prions if the divide-and-conquer approach imposed by limitations in current structural approaches is valid. Or in other words: can the assembly of full-length prions simply be derived from the sum of the two folds observed for the isolated domains?
What can SSNMR do for the Study of Prions?
Solid-state NMR is the method of choice to investigate insoluble non-crystalline biopolymers at atomic resolution. The spectroscopic strategies applied to fibrils are primarily dictated by the linewidth in the sample. The NMR linewidth in the 13C and 15N NMR spectra of prion fibrils and fragments thereof was found to be quite variable. For example, the isolated prion domain of the HET-s protein, prepared under physiological pH conditions, shows lines as narrow as typically observed for crystalline protein preparations (0.1 ppm for the narrowest signalsCitation31). For fibrils of the same protein, grown at pH3, the resolution in the 2D spectra is considerably lowerCitation32 and shows similar linewidths as spectra of the fibrillar form of other short prion fragments investigated. Indeed, the Ure2p prion domain in isolationCitation6,Citation19 as well as fragments of Rnq1pCitation20 and Sup35pCitation22 typically show line widths of 1.5 to 2.5 ppm. The reason for the broadening is not entirely clear, and two effects can play a role: (i) the existence of several well-defined polymorphic fibrils in the sample leads to the superposition of their spectra. Due to the similarity of the polymorphs, their isotropic chemical shifts can be quite similar which leads to a considerable number of non-resolved lines that can result in a broadening of the lines. (ii) The presence of disorder, like the somewhat irregular packing of protofibrils into fibrils, or a variable mass-per-length along a single fibril, or a large number of packing errors due to only small energy differences, e.g., if the register between β-sheets changes. The distinction between polymorphism and a large number of packing errors and minor structural variations is not always straightforward. For the pH3 fibrils of HET-s(218-289), the spectra are compatible with the superposition of a finite number of signals,Citation32 which hints that the explanation (i) could be the correct one in this case. For the isolated Ure2 prion domain, the second explanation could be more probable. In this context, it should be pointed out that there is ample evidence for the existence of different forms of fibrils, which should undoubtedly be classified as polymorphs.Citation33–Citation36 These polymorphs can have significantly different packing schemes as exemplified in the structural model for Aβ by Tycko and coworkers.Citation35 But even in such a case, each of these polymorphs can still be characterized by relatively broad resonance lines, which is an indication that further polymorphism or structural disorder within a polymorph is involved. Indications for the existence of a large number of variations, or polymorphs, comes also from other biophysical methods, e.g., for cryo-electro microscopy.Citation37,Citation38
While broad lines arise from local disorder, the observation of one set of narrow resonances for a fibrillar protein provides in a very sensitive manner evidence for a high local order and structural homogeneity, as for example observed in protein crystals. It also implies for multimeric proteins that all protein-protein interfaces between monomers must be identical or very similar; in crystals with several molecules per unit cell, resonance doubling can often be observed for conformational differences between the monomers, or residues experiencing different crystal contacts.
Resolution is Key
NMR of fibrils that exhibit significant disorder, as judged from the heterogeneous line broadening in NMR spectra, often only allow the measurement of a limited number of distances between selective labels, which then leads to the establishment of structural models satisfying these restraints. In cases where the local order is high enough such that they yield narrow lines under magic-angle spinning conditionsCitation6,Citation24,Citation31,Citation34,Citation39 (e.g., HET-s(218-289),Citation31 Ure2p,Citation6 or the Y145Stop variant or PrPCitation39), structural studies of fibrils have many common elements with solid-state NMR high-resolution 3D structure determination of crystalline proteins,Citation40–Citation45 and basically the same strategies can be applied.Citation40,Citation43–Citation48 They include, as in solution, sequential resonance assignments, followed by the collection of a large set of distance restraints used in structure calculations. This has recently been demonstrated for small crystalline proteinsCitation40,Citation43–Citation45,Citation47 as well as fragments of prion fibrils.Citation24,Citation49 This progress has been made possible due to the availability of higher fields, methodological developments, as well as progress in sample preparation. For full structure determination of larger systems, like full-length prions,Citation6 further developments, including 3D methods, are currently under way.
Sample Preparation for NMR Studies
NMR needs relatively large amounts of bulk sample, preferably several tens of milligrams. For atomic-resolution structure determination, the samples must be isotopically labeled. While small prion peptides as Ure2p(10-39) have been prepared by automated solid-phase synthesis, larger fragments or full-length proteins are usually prepared by expression in E. coli, analogous to the expression of globular proteins. This procedure allows a cost-effective isotopic labeling with 13C, 15N and, in some cases, 2H. Proteins can be expressed in inclusion bodies or can be purified from the soluble fraction. Fibrillization is induced e.g., by a change in concentration, temperature,Citation50 buffer or pH.Citation21 The conditions during expression, purification and fibrillization can be critical for the structure to be obtained. Care has to be taken to fibrillize the protein from its native form, and not from partially folded intermediates, as often encountered in the presence of denaturants. Seeding a certain polymorph or—for prions—with a certain strain—can lead to a fibrillization in the same polymorph as the seed.Citation33 Thus, impurities, as for example cleavage products including isolated prion domains, should be absent, since they might seed non-native fibrils. For multidomain proteins, the entire folding process can be critical to obtain the correctly folded species.Citation6,Citation51,Citation52 Fibrillization yields a gel-like pellet, which can be centrifuged into an MAS rotor. To obtain an optimized signal-to-noise ratio, it is important to pack the rotors as densely as possible. However, extensive dehydration of the sample must be avoided as this leads to significant line-broadening. In our hands, filling the rotors using a centrifuge or an ultracentrifuge turns out to be the easiest and most gentle method.Citation53
HET-s and Ure2p: Structural Models of the Full-Length Prions
Fibrils from HET-s and Ure2p full-length prions have been studied by solid-state NMR. Their molecular size, of 289 and 354 amino-acid residues respectively, is quite large for present solid-state NMR techniques. Nevertheless, high-resolution 2D and 3D spectra of good quality have been obtainedCitation6,Citation7 and a considerable amount of structural information becomes available.
As mentioned above, both proteins show the same organization, which consists of a globular domain and a prion domain. For HET-s, the prion domain is C-terminal, while for Ure2p it is found at the N-terminal part. Both globular domains show a mainly α-helical fold. In contrast to HET-s, which prion domain shows an amino-acid composition that is not different from typical globular proteins, the Ure2p prion domain is rich in N, Q, S and T residues.
The studies of the two proteins followed a similar strategy: investigation of the isolated N- and C-terminal parts, followed by comparison to the full-length protein. The fibrils formed by the fragment HET-s(218-289) corresponding to the prion domain are well ordered on a molecular level and lead to highly resolved spectra, and an initial atomic-resolution structure has been determined from 134 spectrally resolved peaks,Citation24 (). A full structure calculation using over 2,600 distance restraints lead to essentially identical results for the core and a further characterization of the C-terminal part (unpublished data). The structure of HET-s(218-289) fibrils consists of four β-strands forming two windings of a β-solenoid. While the fibrils from the isolated HET-s prion domain reveal a high structural order, the fibrils formed by the isolated Ure2p prion domain reveal significant static disorder.Citation6,Citation19 A comparison of two-dimensional DARR spectra of the two compounds under similar conditions is shown in . A detailed structural analysis of Ure2p(1-93) is thus difficult. Amino-acid specific assignments reveal that the observed chemical shifts mainly correspond to β-sheet secondary structure (as also observed by x-ray diffraction for these amyloid fibrils),Citation54 with some few residues showing dihedral angles typical for turns. Spectra from the isolated globular parts were recorded on crystalline samples and reveal narrow lines for both Ure2p(70-354)Citation6 und HET-s(1-223)Citation7 of the type typically observed in microcrystals.
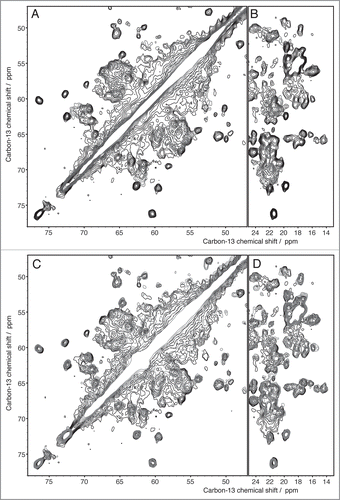
The spectra recorded from the fibrils formed by the full-length proteins yielded unexpected results for both HET-s and Ure2p. For HET-s, the spectra reveal both narrow and broad signals, where the narrow ones show a total conservation of the line shapes and positions when compared to the isolated prion domain. Some extracts of the two-dimensional 13C correlation Spectrum of HET-s are shown (in black contours), and compared with the one of the fragment 218–289 (blue contours), in . All sharp lines in the full-length HET-s spectra can be identified with resonances from the prion-forming domain (residues 218–289) demonstrating that the structure of the amyloid core, as determined for the isolated domain, is fully conserved in the full-length fibrils. Remaining signals, stemming from the globular domain, show broad features and only poorly conserved resonances when compared to the spectra from crystalline globular domain, indicating partial disorder. The spectra are less resolved, less dispersed and less intense than expected indicating that this domain is dynamically disordered; its structure seems at least partially compromised. Even if the chemical shifts correspond still mainly to α-helical conformation, the domain does not show a well-defined structure anymore, and it might be described as a molten globule.Citation7 In fact, the formation of the amyloid backbone seems to force the globular domain into a less ordered state. A linker region can be identified which might connect the globular part to the amyloid core. This structure differs from the model proposed earlier for HET-s,Citation9 but is reminiscent of a model proposed for another prion, Ure2p, where an ordered amyloid core is decorated by disordered globular appendages,Citation9,Citation55 and is shown in . Spectra of Ure2p are quite different and indicative of a different molecular organization. In contrast to HET-s, the two-dimensional spectra are highly resolved, and reveal a well-ordered and rigid globular domain, with one set of resonances observed for isolated resonances. The DARR spectrum is shown in black in . The resonances in this spectrum show similar chemical shifts and linewidths as observed for the isolated globular domain, as assessed by comparison between spectra from the globular domain in crystals (shown in red) and spectra from the full-length protein (in black). Most well resolved resonances can be identified with corresponding resonances in the isolated globular domain of Ure2p, Ure2p(93-354). Thus, the fold of the globular domain is highly conserved in the fibrils, and retains high order, comparable to the one observed in crystals, adding to the evidence that the Ure2p prion is build according to a different structural scheme.Citation6,Citation52,Citation56 Signals originating from the prion domain are not so easily identified in the 2D spectra; there are, however, experimental indications that this part of the molecule is still at least partly disordered in the full-length fibrils, and it is not clear today to what extent chemical shifts and line-widths are conserved. It is evident however that it does not entirely become as highly ordered as the prion domain of the HET-s protein, even if some narrow signals, which do not seem to arise from the globular domain, are observed. Further analyses, including full resonance assignments by means of 3D spectroscopy, currently under progress in our laboratories, are necessary to propose an atomic-resolution model for Ure2p fibrils.
NMR has, in summary, demonstrated that it is well en route to determine the atomic-resolution structure of full-length prions, at least for the parts that are well ordered. So far, it looks like that only one of the domains, either the globular one, as in Ure2p, or the prion domain, as in HET-s, are well-ordered in the context of the full-length fibrils. The comparions of HET-s and Ure2p suggests that there is considerable structural diversity in the fibrils of globular domain-containing prions, despite their similar appearance at the microscopic level.
Figures and Tables
Figure 1 Prions identified today and characterized as consisting of a prion domain (blue) and a globular domain (red).
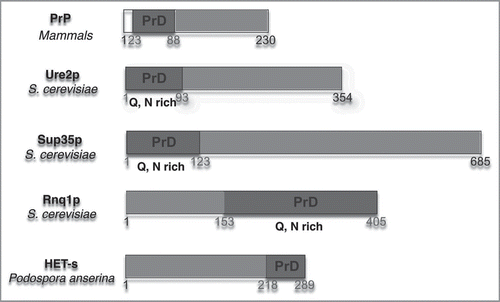
Figure 2 Structure of HET-s (A) five moneomers out of a HET-s(218-289) protofilbril. One monomers forms two turns of a β-solenoid. (B) NMR bundle: superposition on residues N226 to G242, N262 to G278 of the 20 lowest-energy structures of a total of 200 calculated HET-s(218-289) structures. Only the central molecule of the heptamer is shown.Citation24
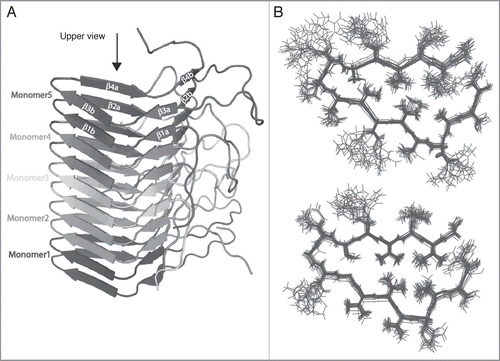
Figure 3 DARR spectra (100 ms and 20 ms mixing, respectively) of the prion domains of HET-s and Ure2p reveal the considerably higher order encountered in HET-s(218-289) if compared to Ure2p(1-93) which represents itself in a narrow linewidth (see main text) HET-s spectrum adapted from,Citation24,Citation31 Ure2p adapted from.Citation6
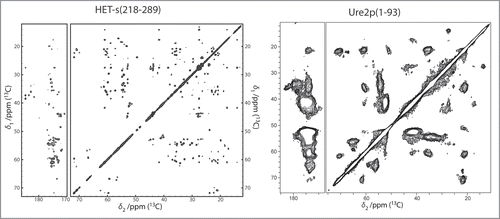
Figure 4 (A and B) Extracts of the 100 ms DARR spectra of HET-s(218-289) (blue) and HET-s(black). All peaks of the prion domain are present in the spectrum of the HET-s fibrils. (C and D) Extracts of the 100 ms DARR spectra of HET-s(1-227) (red) and HET-s(black). The structure of the prion domain is the same as in HET-s(218-289) while the globular domain looses its well-defined teritiary structure.Citation7
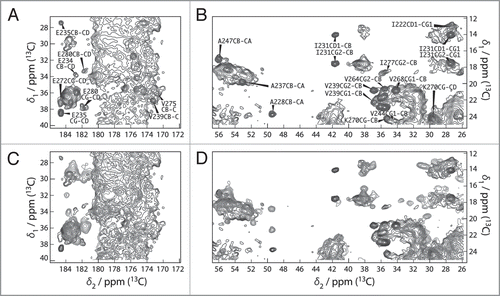
Figure 5 Structural model of full-length HET-s. A Side view and B top view of 10 HET-s molecules within a HET-s amyloid fibril. The ellipsoids represent the N-terminal domains (residues 1–217) which structure is not precisely known in this context. Each molecule is colored uniquely.Citation7
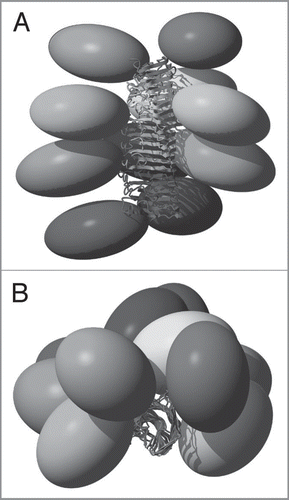
Figure 6 Extracts from 2D 13C DARR spectra recorded with 20 ms mixing time of Ure2p (black), Ure2p1-93 (blue) and Ure2p70-354 (red) (A and B) indicate that the prion domain is structurally different in its isolated form than in the context of the full-length protein. The globular domain, in contrast, is clearly preserved, as seen in (C and D).
Acknowledgements
Many discussions with present and former members of our groups are acknowledged as well as funding by the Agence Nationale de la Recherche (ANR-JC05_44957, ANR-07-PCVI-0013-03, ANR-06-BLAN-0266, ANR-PCV08_321323 and ANR08-PCVI-0022-02), the ETH Zurich, the ETHIRA grant system and the Swiss National Science Foundation. We acknowledge a Germaine de Staël stipend for the collaboration between the labs involved.
References
- Legname G, Nguyen HB, Peretz D, Cohen FE, DeArmond SJ, Prusiner SB. Continuum of prion protein structures enciphers a multitude of prion isolate-specified phenotypes. Proc Natl Acad Sci USA 2006; 103:19105 - 19110
- Cohen FE, Prusiner SB. Pathologic conformations of prion proteins. Annual Review of Biochemistry 1998; 67:793
- Prusiner SB. Novel Proteinaceous Infectious Particles Cause Scrapie. Science 1982; 216:136 - 144
- Tanaka M, Chien P, Naber N, Cooke R, Weissman JS. Conformational variations in an infectious protein determine prion strain differences. Nature 2004; 428:323 - 328
- Toyama BH, Kelly MJS, Gross JD, Weissman JS. The structural basis of yeast prion strain variants. Nature 2007; 449:233 - 238
- Loquet A, Bousset L, Gardiennet C, Sourigues Y, Wasmer C, Habenstein B, et al. Prion Fibrils of Ure2p Assembled under Physiological Conditions Contain Highly Ordered, Natively Folded Modules. J Mol Biol 2009; 394:108 - 118
- Wasmer C, Schütz A, Loquet A, Buhtz C, Greenwald J, Riek R, et al. The Molecular Organization of the Fungal Prion HET-s in Its Amyloid Form. J Mol Biol 2009; 394:119 - 127
- Wickner RB, Taylor KL, Edskes HK, Maddelein M, Moriyama H, Roberts BT. Prions in Saccharomyces and Podospora spp.: Protein-Based Inheritance. Microbiol Mol Biol Rev 1999; 63:844 - 861
- Wickner RB, Edskes H, Shewmaker F, Nakayashiki T. Prions of fungi: inherited structures and biological roles. Nat Rev Microbiol 2007; 5:611 - 618
- Wickner RB. [URE3] as an altered URE2 protein: evidence for a prion analog in Saccharomyces cerevisiae. Science 1994; 264:566 - 569
- Patel BK, Liebman SW. “Prion-proof” for [PIN+]: infection with in vitro-made amyloid aggregates of Rnq1p-(132-405) induces [PIN+]. J Mol Biol 2007; 365:773 - 782
- Patino MM, Liu JJ, Glover JR, Lindquist S. Support for the prion hypothesis for inheritance of a phenotypic trait in yeast. Science 1996; 273:622 - 626
- Du Z, Park K-W, Yu H, Fan Q, Li L. Newly identified prion linked to the chromatin-remodeling factor Swi1 in Saccharomyces cerevisiae. Nat Genet 2008; 40:460 - 465
- Patel BK, Gavin-Smyth J, Liebman SW. The yeast global transcriptional co-repressor protein Cyc8 can propagate as a prion. Nat Cell Biol 2009; 11:344 - 349
- Zahn R, Liu A, Luhrs T, Riek R, von Schroetter C, Lopez Garcia F, et al. NMR solution structure of the human prion protein. Proc Natl Acad Sci USA 2000; 97:145 - 150
- Riek R, Hornemann S, Wider G, Billeter M, Glockshuber R, Wuthrich K. NMR structure of the mouse prion protein domain PrP(121-231). Nature 1996; 382:180 - 182
- Balguerie A, Dos Reis S, Ritter C, Chaignepain S, Coulary-Salin B, Forge V, et al. Domain organization and structure-function relationship of the HET-s prion protein of Podospora anserina. EMBO J 2003; 22:2071 - 2081
- Pierce M, Baxa U, Steven A, Bax A, Wickner R. Is the prion domain of soluble Ure 2p unstructured?. Biochemistry 2005; 44:321 - 328
- Baxa U, Wickner RB, Steven AC, Anderson DE, Marekov LN, Yau W, et al. Characterization of beta-sheet structure in Ure2p(1-89) yeast prion fibrils by solid-state nuclear magnetic resonance. Biochemistry 2007; 46:13149 - 13162
- Wickner RB, Dyna F, Tycko R. Amyloid of Rnq1p, the basis of the [PIN+] prion, has a parallel in-register b-sheet structure. Proc Natl Acad Sci USA 2008; 105:2403 - 2408
- Ritter C, Maddelein ML, Siemer AB, Luhrs T, Ernst M, Meier BH, et al. Correlation of structural elements and infectivity of the HET-s prion. Nature 2005; 435:844 - 848
- Shewmaker F, Wickner RB, Tycko R. Amyloid of the prion domain of Sup35p has an in-register parallel beta-sheet structure. Proc Natl Acad Sci USA 2006; 103:19754 - 19759
- Nelson R, Sawaya MR, Balbirnie M, Madsen AO, Riekel C, Grothe R, et al. Structure of the cross-beta spine of amyloid-like fibrils. Nature 2005; 435:773 - 778
- Wasmer C, Lange A, van Melckebeke H, Siemer AB, Riek R, Meier BH. Amyloid Fibrils of the HET-s(218-289) Prion Form a {beta} Solenoid with a Triangular Hydrophobic Core. Science 2008; 319:1523 - 1526
- Bousset L, Belrhali H, Melki R, Morera S. Crystal structures of the yeast prion Ure2p functional region in complex with glutathione and related compounds. Biochemistry 2001; 40:13564 - 13573
- Umland TC, Taylor KL, Rhee S, Wickner RB, Davies DR. The crystal structure of the nitrogen regulation fragment of the yeast prion protein Ure2p. Proc Natl Acad Sci USA 2001; 98:1459 - 1464
- Greenwald J, Buhtz C, Ritter C, Kwiatkowski W, Choe S, Maddelein M-L, et al. The mechanism of prion inhibition by HET-S 2010; (submitted for publication)
- Lysek DA, Schorn C, Nivon LG, Esteve-Moya V, Christen B, Calzolai L, et al. Prion protein NMR structures of cats, dogs, pigs and sheep. Proc Natl Acad Sci USA 2005; 102:640 - 645
- Gossert AD, Bonjour S, Lysek DA, Fiorito F, Wuthrich K. Prion protein NMR structures of elk and of mouse/elk hybrids. Proc Natl Acad Sci USA 2005; 102:646 - 650
- Zahn R, Guntert P, von Schroetter C, Wuthrich K. NMR Structure of a Variant Human Prion Protein with Two Disulfide Bridges. J Mol Biol 2003; 326:225 - 234
- Siemer AB, Ritter C, Ernst M, Riek R, Meier BH. High-resolution solid-state NMR spectroscopy of the prion protein HET-s in its amyloid conformation. Angew Chem Int Ed 2005; 44:2441 - 2444
- Wasmer C, Soragni A, Sabaté R, Lange A, Riek R, Meier BH. Infectious and noninfectious amyloids of the HET-s(218-289) prion have different NMR spectra. Angew Chem Int Ed 2008; 47:5839 - 5841
- Petkova AT, Leapman RD, Guo Z, Yau W, Mattson MP, Tycko R. Self-Propagating, Molecular-Level Polymorphism in Alzheimer's {beta}-Amyloid Fibrils. Science 2005; 307:262 - 265
- Heise H, Hoyer W, Becker S, Andronesi OC, Riedel D, Baldus M. Molecular-level secondary structure, polymorphism, and dynamics of full-length {alpha}-synuclein fibrils studied by solid-state NMR. Proc Natl Acad Sci USA 2005; 102:15871 - 15876
- Paravastu AK, Leapman RD, Yau W, Tycko R. Molecular structural basis for polymorphism in Alzheimer's beta-amyloid fibrils. Proc Natl Acad Sci USA 2008; 105:18349 - 15354
- Paravastu AK, Petkova AT, Tycko R. Polymorphic Fibril Formation by Residues 10–40 of the Alzheimer's {beta}-Amyloid Peptide. Biophys J 2006; 90:4618 - 4629
- Fändrich M, Meinhardt J, Grigorieff N. Structural polymorphism of Alzheimer Abeta and other amyloid fibrils. Prion 2009; 3:89 - 93
- Meinhardt J, Sachse C, Hortschansky P, Grigorieff N, Fändrich M. Abeta(1–40) fibril polymorphism implies diverse interaction patterns in amyloid fibrils. J Mol Biol 2009; 386:869 - 877
- Helmus JJ, Surewicz K, Nadaud PS, Surewicz WK, Jaroniec CP. Molecular conformation and dynamics of the Y145Stop variant of human prion protein in amyloid fibrils. Proceedings of the National Academy of Sciences 2008; 105:6284 - 6289
- Castellani F, van Rossum B, Diehl A, Schubert M, Rehbein K, Oschkinat H. Structure of a protein determined by solid-state magic-angle-spinning NMR spectroscopy. Nature 2002; 420:98 - 102
- Castellani F, van Rossum BJ, Diehl A, Rehbein K, Oschkinat H. Determination of solid-state NMR structures of proteins by means of three-dimensional N-15-C-13-C-13 dipolar correlation spectroscopy and chemical shift analysis. Biochemistry 2003; 42:11476 - 11483
- Böckmann A. High-resolution solid-state MAS NMR of proteins-Crh as an example. Magn Reson Chem 2007; 45:24 - 31
- Loquet A, Bardiaux B, Gardiennet C, Blanchet C, Baldus M, Nilges M, et al. 3D Structure Determination of the Crh Protein from Highly Ambiguous Solid-State NMR Restraints. J Am Chem Soc 2008; 130:3579 - 3589
- Manolikas T, Herrmann T, Meier BH. Protein structure determination from C-13 spin-diffusion solid-state NMR spectroscopy. J Am Chem Soc 2008; 130:3959 - 3966
- Franks WT, Wylie BJ, Frericks Schmidt HL, Nieuwkoop AJ, Mayrhofer R, Shah GJ, et al. Dipole tensor-based atomic-resolution structure determination of a nanocrystalline protein by solid-state NMR. Proceedings of the National Academy of Sciences 2008; 105:4621 - 4624
- Seidel K, Etzkorn M, Heise H, Becker S, Baldus M. High-Resolution Solid-State NMR Studies on Uniformly 13C,15N-Labeled Ubiquitin. ChemBioChem 2005; 6:1638 - 1647
- Zech SG, Wand AJ, McDermott AE. Protein structure determination by high-resolution solid-state NMR spectroscopy: Application to microcrystalline ubiquitin. J Am Chem Soc 2005; 127:8618 - 8626
- Korukottu J, Schneider R, Vijayan V, Lange A, Pongs O, Becker S, et al. High-resolution 3D structure determination of kaliotoxin by solid-state NMR spectroscopy. PLoS ONE 2008; 3:2359
- Siemer AB, Ritter C, Steinmetz MO, Ernst M, Riek R, Meier BH. 13C, 15N Resonance Assignment of Parts of the HET-s Prion Protein in its Amyloid Form. J Biomol NMR 2006; 34:75 - 87
- Kammerer RA, Kostrewa D, Zurdo J, Detken A, Garcia-Echeverria C, Green JD, et al. Exploring amyloid formation by a de novo design. Proc Natl Acad Sci USA 2004; 101:4435 - 4440
- Thual C, Bousset L, Komar AA, Walter S, Buchner J, Cullin C, et al. Stability, folding, dimerization and assembly properties of the yeast prion Ure2p. Biochemistry 2001; 40:1764 - 1773
- Bousset L, Thomson NH, Radford SE, Melki R. The yeast prion Ure2p retains its native alpha-helical conformation upon assembly into protein fibrils in vitro. EMBO J 2002; 21:2903 - 2911
- Böckmann A, Gardiennet C, Verel R, Hunkeler A, Loquet A, Pintacuda G, et al. Characterization of different water pools in solid-state NMR protein samples. J Biomol NMR 2009; 45:319 - 327
- Baxa U, Cheng N, Winkler D, Chiu T, Davies D, Sharma D, et al. Filaments of the Ure2p prion protein have a cross-β core structure. J Struct Biol 2005; 150:170 - 179
- Kajava AV, Baxa U, Wickner RB, Steven AC. A model for Ure2p prion filaments and other amyloids: The parallel superpleated {beta}-structure. Proc Natl Acad Sci USA 2004; 101:7885 - 90
- Bousset L, Briki F, Doucet J, Melki R. The native-like conformation of Ure2p in fibrils assembled under physiologically relevant conditions switches to an amyloid-like conformation upon heat-treatment of the fibrils. J Struct Biol 2003; 141:132 - 42