Abstract
Prion diseases are fatal neurodegenerative disorders caused by prion proteins (PrP). Infectious prions accumulate in brain through a template mediated conformational conversion of endogenous PrPC into alternately folded PrPSc. Immunoassays toward pre-clinical detection of infectious PrPSc have been confounded by low-level prion accumulation in non-neuronal tissue and the lack of PrPSc selective antibodies. We report a method to purify infectious PrPSc from biological tissues for use as an immunogen and sample enrichment for increased immunoassay sensitivity. Significant prion enrichment is accomplished by sucrose gradient centrifugation of infected tissue and isolation with detergent resistant membranes from lipid rafts (DRMs). At equivalent protein concentration a 50-fold increase in detectable PrPSc was observed in DRM fractions relative to crude brain by direct ELISA. Sequential purification steps results in increased specific infectivity (DRM >20-fold and purified DRM immunogen >40-fold) relative to 1% crude brain homogenate. Purification of PrPSc from DRM was accomplished using phosphotungstic acid protein precipitation after proteinase-K (PK) digestion followed by size exclusion chromatography to separate PK and residual protein fragments from larger prion aggregates. Immunization with purified PrPSc antigen was performed using wild-type (wt) and Prnp0/0 mice, both on Balb/cJ background. A robust immune response against PrPSc was observed in all inoculated Prnp0/0 mice resulting in antisera containing high-titer antibodies against prion protein. Antisera from these mice recognized both PrPC and PrPSc, while binding to other brain-derived protein was not observed. In contrast, the PrPSc inoculum was non-immunogenic in wt mice and antisera showed no reactivity with PrP or any other protein.
Introduction
Prion diseases are a family of progressive, fatal neurodegenerative disorders caused by the accumulation of the alternatively folded prion protein PrPSc. In the CNS, prions produce neuronal cell death, spongiform vacuolation and gliosis.Citation1 The PrPSc protein is extractable from diseased tissue and biochemically distinguished from endogenous PrPC by partial protease resistance and detergent insolubility.Citation2 Both PrPC and PrPSc share the same amino acid sequence, but PrPSc adopts an abnormal conformation that is transmissible and serves as a template for the conversion of host PrPC into the pathogenic prion isoform.Citation3,Citation4 The mechanism responsible for the transmission, conformational conversion of PrPC to PrPSc and subsequent disease progression remains enigmatic.
Detection of infectious prions relies on combined use of immunoassay and histopathological assessment of brain tissue from infected animals.Citation5 Current immunoassays are dependent on antibodies that recognize both the normal and abnormal isoforms of PrP. To distinguish abnormal PrPSc from normal PrPC requires limited digestion with proteinase-K (PK) to hydrolyze PK-sensitive PrPC while retaining the PK-resistant PrPSc (PrP 27–30). The PrP 27–30 protein is smaller than PrPC and intact PrPSc and thus can be recognized by a mobility shift following SDS-PAGE and Western blot detection with anti-PrP antibodies.Citation6,Citation7 This methodology is effective for the identification of PrPSc from prion enriched samples such as brain. Yet prion accumulation in the brain is progressive and infected asymptomatic animals pose significant sampling challenges. Indeed, there appears to be minimal shedding or accumulation of PrPSc in other more accessible tissue or fluid compartments.Citation8,Citation9 Moreover, variability in the efficacy of prion proteolysis of samples confounds detection of low-level PrPSc.Citation10 Definitive assessment of prion infected animals is determined in post-mortem evaluation of brain tissue by immunoassay and histopathology from clinically symptomatic animals.
There remains an acute need for a sensitive and selective prion immunodiagnostic assay capable of pre-clinical assessment of infected animals from accessible tissues or fluids.Citation11 Most immunoassay detection limits are insufficient to detect low-level prion contamination that can transmit disease by bioassay. Current assays are confounded by reliance on removal of PK-sensitive PrPC as no antibody has emerged that can selectively distinguish infectious PrPSc from PrPC.Citation12 The need to remove PrPC protein from samples often diminishes immunoassay sensitivity by reducing the amount of PrPSc and increasing assay background. Moreover, the occurrence of PK-sensitive PrPSc isoforms poses additional concerns for many immunodiagnostic assays.Citation13
The difficulty of prion antibody generation is underscored by the identical primary structure of normal and abnormal PrP protein isoforms and isolation of purified infectious prion. The use of synthetic PrP peptides or recombinant PrPC has been successful in generating anti-PrP antibodies for detection of both PrPC and PrPSc proteins.Citation14,Citation15 However, PrPC antigen has proven to be a poor immunogen as endogenous PrPC protein negates a robust immune response.Citation16,Citation17 The immunogenicity of PrPC antigen has been improved by using Prnp0/0 mice with resulting production of high-affinity anti-PrP antibodies.Citation14 However, the use of a PrPC antigen invariably leads to production of antibodies that recognize PrPC with a low probability of generating a PrPSc selective antibody. Here we report a simple methodology for the enrichment and purification of infectious PrPSc antigen from brain for immunization of Prnp0/0/Balb/cJ mice. We show that purified PrPSc antigen elicits a robust immune response in Prnp0/0, but not wt Balb/cJ, mice resulting in the production of antisera with significant anti-PrP activity. Notably, no antibodies were generated against other brain proteins confirming the relative purity and utility of the PrPSc antigen preparation.
Results
A sucrose density gradient effectively separates soluble proteins from those that are detergent resistant at 4°C by exploiting differences in buoyancy properties due to lipid-association of some proteins. A measure of total protein following fractionation of crude brain homogenate (12 × 1 mL) showed that the majority of proteins (>99%) were retained at the bottom of the sucrose gradient between fractions #8–12 (40% sucrose), whereas a small protein peak (<1%) migrated to the zone of the 5/30% sucrose in fractions #4–5 corresponding to detergent resistant membrane (DRM) derived from lipid rafts (). Western blots of the gradient fractions from normal and diseased hamster brain reveal that most of the PrPC (; top left part) and PrPSc (; top right part) were found in the DRM fraction #4 of the minor protein peak. As a positive control for DRM proteins, Flotillin-1, a known lipid raft protein, was detected by Western blotting from the same gradient fractions and localized as expected to DRM fraction #4 (; bottom parts). Aliquots of each gradient fraction were used as antigen in a direct binding ELISA for detection of PrP protein. Confirming the Western blots, the majority of PrPC from normal brain (; left part) and PrPSc from diseased brain (; right part) were localized in the DRM-enriched fraction #4.
Prion enrichment was evaluated by comparing levels of PK-resistant PrPSc in crude brain homogenate to DRM gradient fractions. At equal concentration of total protein the DRM fraction contained ∼50-fold more detectable prion relative to crude brain homogenate by direct ELISA (; t-test p < 0.001). Prion enrichment in DRM was further evaluated by Western blot with a detectable increase in band intensity for all three PrPSc glycoforms in DRM fractions relative to crude brain homogenates (). PTA precipitation of DRM fractions resulted in increased prion detection relative to the DRM fraction alone.
compares the oligomeric status of the PrP found in normal and scrapie enriched DRM preparations from brain homogenates. The DRM fractions were subjected to sedimentation gradient centrifugation in a linear sucrose gradient. Each gradient was then fractionated into 12 equal fractions and each fraction evaluated by Western blot probing with anti-PrP antibody (; top part), anti-Caveolin-1 antibody (; middle part) and anti-Flotillin-1 antibody (; bottom part). PrPC in the DRM from normal brain resides as monomeric to high molecular weight aggregate with the detection of mono- and di-glycosylated isoforms predominant (; top left part). In contrast, the PrPSc in the DRM from diseased brain was observed as high molecular weight aggregate >400 kDa dominated by the mono- and di-glycosylated isoforms (; top right part). Moreover, non-glycosylated PrP was readily detectable across the gradient at varied molecular weights in DRMs from prion infected brain, yet this isoform was PK-sensitive (; bottom right part) suggesting detection of PrPC. Caveolin-1 and Flotillin-1, established lipid raft proteins,Citation18 were used as markers to validate the integrity of the gradients and were detected as high molecular weight oligomers (; bottom left parts).
Limited PK-digestion of PrPSc positive DRM fractions resulted in the proteolytic degradation of PrPC and the N-terminal truncation of PrPSc to form PrP 27–30. Importantly, the PK-digested samples also included peptide fragments, PK and lipids. To isolate PrPSc from these contaminants, we exploited preferential binding and precipitation of PrPSc by PTA. Following PK-digestion of PrPSc DRM fraction, proteins were PTA precipitated then solubilized and fractionated over Sephadex G100. A major protein peak emerged at the column void fraction #5 (void >100 kDa; ; top part; solid line) where PrPSc was detected by ELISA (; top part; dashed line). Evaluation of eluted G100 fractions by western blot shows that the majority of PrPSc was detected in the void fraction #5 (). shows that PrPSc was further concentrated by a second PTA precipitation after elution from G100 (). Electrophoresis of the PTA concentrated G100 eluted fraction #5 followed by silver (; left part) and Coomassie blue staining (; middle part) revealed proteins at the correct molecular mass corresponding to PrPSc detected by western blot (; right part).
Using the hamster bioassay we showed that the DRM fraction (PrPSc DRM) isolated from prion-infected brain homogenate remained infectious (). Likewise, the purified PK-resistant PrPSc from DRM fractions that had been PTA precipitated and fractionated over Sephadex G100 (PrPSc DRM-PK-PTA-G100) retained infectivity (). Our purification scheme resulted in a progressive increase in both the ID50 and a >20-fold and >40-fold increase in specific infectivity of DRM fractions and purified DRM (PrPSc DRM-PK-PTA-G100) relative to 1% crude brain homogenate, respectively ().
Prnp0/0/Balb/cJ mice were immunized with a PrPSc fraction enriched for prion infectivity according to the purification scheme outlined in . Antisera from PrPSc immunized Prnp0/0/Balb/cJ mice (#1–2) bound recombinant Syrian hamster (recSHa) PrP(90-231) at an end-point dilution >1:30,000 by ELISA (). No SHa PrP(90-231) binding was observed using sera from immunized wt Balb/cJ mice (#3–4). Western slot-blot experiments compared binding of antisera from immunized Prnp0/0/Balb/cJ mice #1–2 and wt Balb/cJ mice #3–4 () to recSHaPrP(90-231), SHaPrPSc DRM without limited PK-digestion (PrPC plus PrPSc) and SHaPrP 27–30 in DRMs. Antisera from immunized Prnp0/0/Balb/cJ mice was effective in detecting both PrPC and PrPSc proteins at dilutions >25 K. In contrast, antisera from immunized wt Balb/cJ mice did not detect PrP proteins at any dilution tested. The lack of immunoreactive sera from immunized wt Balb/cJ mice to other DRM proteins further supports the purity of the prion inoculum and lack of immunogenicity of the PrPSc protein in wt animals.
Further characterization of antisera binding to protein in crude brain homogenate and brain-derived DRM fractions from normal (PrPC) and disease (PrPSc) hamster brain with (+) and without (−) PK-digestion is shown from a representative immunized Prnp0/0/Balb/cJ mouse by Western blot (; top part). Weak binding to PrPC was observed in normal brain homogenate (30 µg/lane) compared to the strong binding to PrPC from enriched DRM (10 µg/lane) preparation (, PrPC; arrow). Limited PK-digestion of normal brain samples resulted in the complete loss of detectable PrPC using the antisera. Strong binding to PrPSc was observed in both crude brain homogenates and DRM preparations from disease brain with detection of PrP 27–30 at the expected molecular mass. Importantly, antisera from immunized Prnp0/0/Balb/cJ mice only detected PrP proteins. As a control, a companion blot was probed with the anti-PrP monoclonal antibody IPC1 (; bottom part). A similar pattern of binding to PrPC and PrPSc was observed with IPC1 as compared to the antisera from immunized Prnp0/0/Balb/cJ mice.
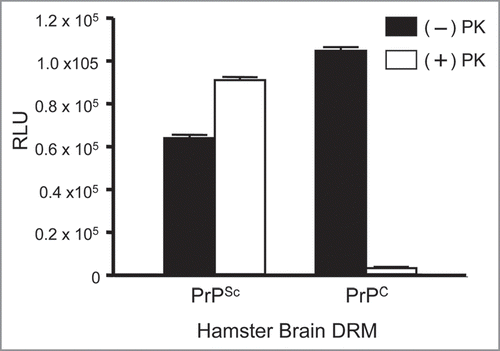
To determine if antisera from prion immunized mice binds to non-denatured prion proteins we performed ELISA on hamster brain DRM fractions (). Our data shows that the antisera was equally effective in the detection of the native prion protein from both normal (PrPC) and diseased (PrPSc) DRM fractions. However, following PK treatment the antisera bound only PK-resistant PrP27–30 from diseased brain (PrPSc) with binding levels equivalent to background in PK-treated normal brain (PrPC). Thus sera from mice immunized with the purified prion preparation have antibodies that recognized both the normal (PrPC) and abnormal (PrPSc) prion isoforms. Importantly, there was no detection of other brain proteins which supports the validity of this strategy for use with hybridoma technology to isolate sensitive and selective anti-prion monoclonal antibodies.
Discussion
The most common methods for the diagnostic confirmation of prion disease involve clinical assessment, followed by postmortem histopathological evaluation of brain tissue along with biochemical detection of PrP 27–30.Citation19,Citation20 Several problems have confounded the pre-clinical diagnostic detection of prion. First, accumulation of PrPSc increases progressively over time; second, most PrPSc resides in the brain which imposes biopsy challenges. Third, prion concentrations below current immunoassay detection limits can transmit disease in animal bioassay.Citation21,Citation22 Fourth, no direct detection method has been developed that can distinguish PrPSc from PrPC without enzymatic or chemical manipulation to render endogenous PrPC undetectable while retaining PrPSc activity. Indeed, no antibody has emerged that can selectively bind PrPSc. Moreover, no surrogate analyte has been identified that can identify prion disease in preclinical animals.Citation23,Citation20 Finally, species and prion strain variability presents additional detection challenges as a result of distinct tissue distribution and availability.Citation24,Citation25
Useful biochemical methods have emerged for the enrichment of PrPSc from brain homogenates that take advantage of differences in sedimentation and solubility.Citation26,Citation27 Yet, these preparative methods have proven insufficient to yield PrPSc enriched fractions suitable for crystal formation or as immunogen for the generation of PrPSc selective antibodies. Several factors likely contribute to the inability to generate a PrPSc selective antibody. First, the choice and preparation of inoculum have favored the generation of PrPC antibodies. The use of recombinant PrPC invariably yields antibodies that recognize PrPC. Moreover, preparation of a native PrPSc is often confounded by contaminating proteins including PrPC. Second, wt animals expressing endogenous PrPC may provide a less robust system for the generation of PrPSc antibodies.Citation28 Third, the method used for screening antibodies requires the selective discrimination of those that bind PrPC from those that bind PrPSc. Here we report the development of prion purification methodology that yields abundant PrPSc from diseased brain () and demonstrate a progressive increase in specific infectivity of prions in DRMs (>20-fold) and the purified DRM-PK-PTA-G100 (>40-fold) inoculum relative to a 1% crude brain homogenate. Moreover, we show that purified PrPSc is highly immunogenic in Prnp0/0/Balb/cJ, but not wt Balb/cJ, mice resulting in generation of high-titer antisera with selective activity to PrP.
Post-translational processing of PrPC includes the addition of a GPI-anchor that targets the protein to lipid rafts of the plasma membrane.Citation29,Citation30 This localization is important for the conformational conversion of PrPC into infectious prion.Citation31–Citation34 Biochemically these lipid-rich membrane domains can be isolated by sucrose density centrifugation as buoyant detergent resistant membranes (DRMs) containing both PrPC and PrPSc proteins.Citation29,Citation10 Exploitation of this methodology allows for a significant enrichment and purification, effectively separating >99% of brain protein from prion in a single step. The resulting DRM fraction is enriched in PrPSc (>40-fold) relative to crude brain homogenate and represents <1% total brain proteins. Prions in the DRM fraction remain soluble and are highly infectious in bioassay. Treatment of DRM fractions with PK effectively removes PrPC and results in PrPSc truncation to form PrP 27–30.
Phosphotungstic acid (PTA) has been used effectively to precipitate prion protein.Citation35,Citation36 The application of PTA to precipitate PrPSc after PK-digestion from DRMs provides an effective method to both concentrate and separate prion from lipids in the enriched DRM preparation. In addition, we found PrPSc in brain DRM fractions is highly aggregated with a molecular mass of >400 kDa. We exploited this property by using size exclusion chromatography to further separate PrPSc from residual PK (<30 kDa) and other protein fragments. Chromatography over Sephadex G100 allowed us to rapidly collect the majority of PrPSc in the column void fraction while residual proteins <100 kDa were retarded by the gel. The resulting PrPSc fraction (PrPSc DRM-PK-PTA-G100) retained infectivity as determined in bioassay. The PrPSc fractions from multiple column separations were pooled and concentrated by a second PTA precipitation to yield a highly purified PrPSc pellet (PrPSc DRMPK-PTA-G100-PTA). This method (outlined in ) allowed for sufficient accumulation of PrPSc devoid of endogenous PrPC and other contaminating proteins to yield a Coomassie stainable band of the correct molecular mass that corresponded to PrPSc by Western blot.
The described methodology resulted in a purified PrPSc antigen of sufficient quantity for multiple immunizations. Since pure PrPSc still shares the same primary amino acid sequence as PrPC, immunizaton with purified prion still presents additional challenges to the immune system to generate a PrPSc selective antibody. There remains a need for the immune system to present a structurally unique PrPSc target in order to generate a selective antibody capable of distinguishing PrPSc from PrPC. Indeed antigen processing of prion by the immune system may likely results in a majority of the prion fragments being recognized as “self ” thus provoking a minimal (or no) immune response in wt animals or production of antibodies that cross-react with PrPC. The structural distinction of PrPSc by the immune system likely represents a statistically rare event. Such an interpretation is supported by its failure to recognize PrPSc as foreign following immunization of wt mice with the purified PrPSc.
The development of a Prnp0/0/Balbc/J mouse provides a useful genetic background to promote a robust immune response to prions and production of monoclonal antibodies. These mice offer a syngenic background for myeloma-spleenocyte fusion and subsequent hybridoma generation. Indeed, our data shows that despite the purity of our prion inoculum it serves as a poor immunogen in wt Balb/cJ mice. However, the lack of detection of any other brain DRM proteins using the antisera from immunized wt Balbc/J mice supports the purity of our prion inoculum. In sharp contrast, immunization of Prnp0/0/Balb/cJ mice elicits a robust immune response that resulted in production of high-titer antiprion specific antiserum. The response appears comparable by Western blot to that observed with established monoclonal anti-PrP antibodies. Additionally, no antibodies against other brain DRM proteins were detected in the antisera from these mice again suggesting a highly purified inoculum.
Antisera from the Prnp0/0/Balb/cJ mice recognized both PK-sensitive and -resistant PrP proteins. This is not surprising given the identical primary amino acid sequence of PrPSc to PrPC and it suggests that a pool of antibodies were produced against multiple epitopes resulting in cross-reactivity with sera by immunoassay. However, the model outlined here is well suited for the generation of hybridomas and isolation of monoclonal antibodies. Efforts are underway to isolate monoclonal antibodies and identify those that bind PrPSc specific epitopes. In conclusion, we have developed a method for the enrichment of infectious prions from lipid rafts and their subsequent purification in sufficient quantity to elicit a robust and selective immune response in Prnp0/0/Balb/cJ mice.
Materials and Methods
All animals were housed in pairs on a 12 h light-dark cycle and provided continual access to food and water. All protocols were approved by the USDA animal care and use committee and experimental procedures conducted in certified BL2 laboratory. Hamster-passaged Sc237 scrapie prions were propagated in female Syrian Golden hamsters (LVG; Charles Rivers Laboratory, MA) beginning at 4 wk of age. Prion-infected hamsters were sacrificed when clinical symptoms included increased startle response, ataxia and >5 s righting reflex. Prnp0/0/Balb/cJ mice were generated at the University of California, San Francisco under approved animal protocols by speed congenic backcrossing 129/SvJ/C57-BL6 Prnp0/0 to inbred Balb/cJ mice and homozygosity verified by PCR as previously described.Citation37,Citation14 Antisera was obtained from anesthetized mice following transcardiac puncture with a 20-gauge needle attached to a 3 mL syringe, transferred to a BD Vacutainer SST tube (BD Biosciences), allowed to clot and sera collected after centrifugation.
Infectious PrPSc was propagated by serial passage in hamster brain following 40 µL intracerebral inoculation of a 1% brain homogenate in 320 mM sucrose using a 27-gauge needle inserted into the right parietal lobe. Detergent resistant membrane (DRM) fractions were diluted in sucrose to a final concentration of 320 mM and inoculated as described. Phosphotungstic acid (PTA) precipitated protein pellets were solubalized in n-octyl-glucoside to final concentration of 60 mM, diluted in sucrose and inoculated as described. Incubation time assay was used to calculate ID50 using the equation Log T = 17 + [Log D] − (0.138*Y) and used for calculation of specific infectivity (ID50/mg inoculum). Onset of clinical scrapie was determined by occurrence of two symptoms in days post-inoculation as defined above. Prnp0/0/Balb/cJ mice starting at 25 d were inoculated (i.p.) with 100 µL antigen using the following regime: two inoculations containing purified PrPSc in RIBI adjuvant (Sigma-Aldrich, MO; Sigma Adjuvant System) separated by 10 d. Sera was collected 3 d after the final inoculation and evaluated for anti-PrP immunoreactivity.
All reagents were of the highest grades commercially available. All antibodies were diluted in 10 mM Tris Buffered Saline with 1% Tween-20 (TBST) containing 1% IgG-free BSA (Jackson Immuno Chemical, PA, USA). Primary antibodies used include: Caveolin-1 rabbit polyclonal diluted 1:1 K (Santa Cruz, CA, USA; N20), Flotillin-1 rabbit polyclonal diluted 1:1 K (Santa Cruz; H-104), IPC1 anti-prion monoclonal diluted 1:10 K (Sigma). Secondary antibodies include: goat-anti-mouse-HRP and goat-antirabbit-HRP diluted 1:10 K (Pierce, IL, USA). Recombinant Syrian hamster (recSHa) PrP(90-231) was generated at UCSF as previously described.Citation38
Hamster brains were homogenized (10% w/v) on ice in 25 mM MES (pH 6.5) with 150 mM NaCL, 1% Triton X-100, 60 mM n-octyl-glucoside, 10 mM PMSF and protease inhibitors (Complete mini; Roche, CH). The homogenate was pre-cleared by centrifugation (1,000x g) at 4°C and supernatant mixed with equal volume of 80% sucrose in 25 mM MES (pH 6.5) with 150 mM NaCL. A 12 mL discontinuous sucrose gradient was formed by applying 4 mL of the 40% brain-sucrose in the bottom of a clear ultra-centrifuge tube (14 × 89 mm; Beckman, CA, USA) followed by a 4 mL layer of 30% MES-Sucrose then 4 mL 5% MES-Sucrose. Tubes were placed in a SW-40T rotor and centrifuged at 39,000 RPM at 4°C for 18 h in a L8-70M class H ultra-centrifuge (Beckman). A visible lipid-rich band corresponding to the detergent resistant membrane (DRM) fraction was observed within the 30–5% sucrose zone and collected (∼1 mL/gradient).
DRM fractions obtained from hamster brain homogenates were mixed with n-octyl-glucoside to a final concentration of 60 mM and incubated at 4°C for 15 min with rotation. A cushion of 250 µL of 50% sucrose in 25 mM MES (pH 6.5) with 150 mM NaCL and 60 mM n-octyl-glucoside was place in the bottom a clear ultra-centrifuge tubes (11 × 60 mm; Beckman) and a 50–5% linear sucrose gradient (4 mL) formed using a mixing gradient maker. The DRM fraction was loaded (300 µL) to the top of the gradient and tubes centrifuged in a SW60 rotor at 50,000 RPM for 10 h at 4°C in a L8-70M class H ultra-centrifuge. 12 × 0.35 mL fractions were collected and analyzed. The gradient was calibrated with known molecular standards as previously described.Citation18
Protein concentration was quantified using a micro-BCA assay (Pierce). Proteinase-K (PK; Roche) treatment was used at a final concentration of 25 µg/mL for brain homogenates and 150 µg/mL for DRM fractions for 1 h at 60°C and inactivation of PK was by denaturation in LDS sample buffer or by addition of PMSF to 10 mM. Electrophoresis was performed on heat denatured samples in LDS buffer normalized by BCA and loaded on 4–12% Bis-Tris gels electrophoresed with MOPS running buffer (Novex; Invitrogen). Gels were transferred to nitrocellulose (Bio-Rad), washed in TBST, blocked with 10% non-fat dry milk, probed with antibodies, protein bands resolved by ECL (Supersignal; Pierce) and imaged on a Flurochem HD documentation system (Alpha Innotech, CA, USA). Gel staining was performed with Coomassie blue (R250; Sigma) or Silver (ProteoSilver Plus; Sigma) and imaged on a light box.
Samples with equivalent protein concentration were diluted in 0.1 M sodium bicarbonate buffer (pH 9.4) and 100 µL absorbed to 96-well maxisorb plates (NUNC, NY) overnight at 4°C. Plates were washed in TBST, blocked in 10% non-fat milk for 1 h at 37°C, incubated 1 h with primary antibody, washed, incubated 1 h with HRP-conjugated secondary antibody, washed, resolved by chemiluminescence (Supersignal; Pierce) detection using Victor2 plate reader (PerkinElmer, MA) and expressed as relative light units (RLU).
A stock of sodium phosphotungstate hydrate (Aldrich, WI, USA) was dissolved at 4% in PBS (pH 7.4) with 170 mM MgCl2. PTA was added to samples to a final concentration of 0.3% with 13 mM MgCL2 and incubated at room temperature for 10 min. Precipitated protein was centrifuged at 10,000x g for 20 min at 4°C, pellets washed repeatedly with 200 mM EDTA in PBS followed by centrifugation with a final wash in ddH20 with remaining water aspirated after centrifugation. PTA pellets were solubalized with n-octyl-glucoside to a final concentration of 60 mM in buffer.
A 15 mL gel bed of Sephadex G100 (Superfine grade, Sigma) was poured in a glass column and equilibrated in 25 mM Tris-HCL (pH 7.4). Column calibration was performed with gel filtration standards (Bio-Rad, CA; #151-1901) and samples loaded at 250 µL in 25 mM Tris-HCL (pH 7.4) with 60 mM n-octyl-glucoside. Proteins were fractionated with 25 mM Tris-HCL (pH 7.4) at a flow rate of 100 µL/min in 1 mL fractions. Column void was defined at 5 mL with detectable high molecular weight standards (>100 kDa) eluted.
Abbreviations
| DRM | = | detergent resistant membranes |
| PK | = | proteinase-K |
| PrPC | = | endogenous PrP protein |
| PrPSc | = | infectious prion protein |
| wt | = | wild-type mice |
| Prnp0/0 | = | Prnp-null mice |
| ELISA | = | enzyme linked immunosorbent assay |
| BL2 | = | biosafety level-2 containment |
| PTA | = | phosphotungstic acid |
| BSA | = | bovine serum albumin |
| ECL | = | enhanced chemiluminescence |
| SHa | = | syrian hamster |
| rec | = | recombinant |
Figures and Tables
Figure 1 Isolation of prions from crude brain homogenate with detergent resistant membranes (DRM). Sucrose density gradient centrifugation separates lipid-associated proteins from the majority of brain proteins via differences in sucrose buoyancy at 4°C. Fractionation of the gradient (12 × 1 mL) resulted in detection of a small protein peak observed within the 5–30% sucrose zone (buoyant fractions #4–5), whereas the bulk of proteins from both PrPC and PrPSc brain homogenate were retained at the bottom of the gradient (A). Western blots revealed that the majority of detectable PrPC and PrPSc were localized to the lipid-rich DRM fractions #4–5 (B; top parts). Proteinase-K (+PK) digestion of the gradient fractions from PrPSc infected brain resulted in an expected molecular weight shift and detection of PrPSc in the DRM fractions (B; top right part). The DRM protein marker Flotillin-1 served as a positive control to confirm the integrity of the gradient and localization of the DRM fractions (B; bottom parts). The majority of PrPC and proteinase-K resistant PrPSc were detected by ELISA in DRM fractions #4–5 (C).
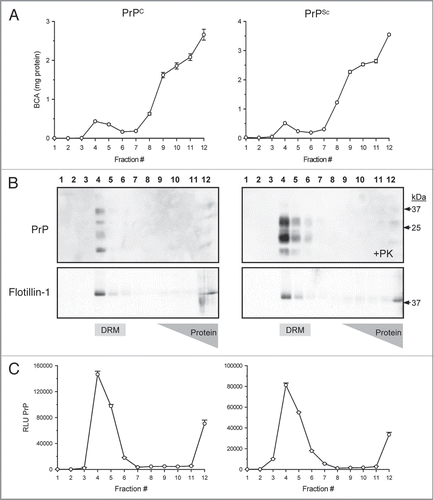
Figure 2 Enrichment of prions in detergent resistant membrane fractions (DRM). Increased detection of PrP 27–30 in DRM fraction (>40-fold) relative to crude brain homogenate by direct ELISA (A). Protein normalization by BCA (5 µg/mL); RLU = relative light units; quantitation of three independent samples (Mean ± SE M; p =<0.001). Western blot comparison of detectable PrP 27–30 in crude hamster brain homogenates, DRM and phosphotungstic acid (PTA) precipitated DRM proteins (B). Progressive increase in detection of three PrPSc glycoforms in DRM fractions and PTA precipitated DRM fractions compared to crude brain homogenates. Protein normalization by BCA prior to PK-treatment and PTA precipitation.
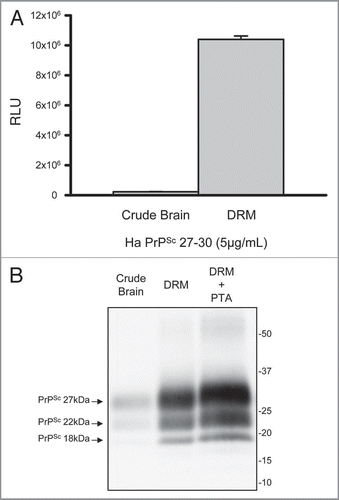
Figure 3 Prion resides as a high molecular weight aggregate in detergent resistant membrane (DRM) fractions. Western blot detection of PrPC following fractionation of normal brain DRM on a linear sucrose gradient showed detection of PrPC across a range of molecular weights composed primarily of mono- and di-glycosylated isoforms (top left part). The oligomeric status of PrPSc from Scrapie infected DRM differs from PrPC with the majority detected as high molecular weight (Fraction #12; >440 kDa) aggregate (top right part). This high molecular weight PrPSc species is composed primarily of mono- and di-glycosylated PrP with detection of non-glycosylated PrP across lower molecular weights (Fractions #3–10). Proteinase-K digestion of gradient fractions from Scrapie DRM shows that all the PK-resistant PrPSc is localized to fraction #12 as a high molecular weight aggregate whereas the non-glycosylated PrP in fractions #3-10 is PK-sensitive (bottom right part). Caveolin-1 (Cav-1) and flotillin-1 (Flot-1) are established DRM protein markers known to form high molecular weight oligomeric complexes and were used as positive controls to validate the integrity of the sucrose gradient (middle and bottom left parts respectively). The linear sucrose gradient was calibrated with know molecular weight protein standards and their mobility and molecular weight are indicated by the large arrows at the bottom of .
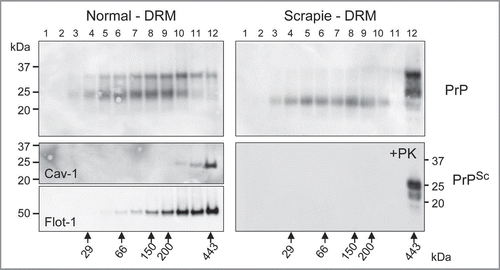
Figure 4 Concentration and purification of prion by PTA and size exclusion chromatography. Enriched PrPSc DRM fractions were treated with proteinase-K then proteins PTA precipitated, the solubalized (PrPSc DRM-PK-PTA) material was then fractionated by size exclusion chromatography (Sephadex G100). Protein concentration of fractions was determined by BCA assay (A; solid line) and PrPSc detection by ELISA (A; dashed line). A small protein peak was observe in fraction #5 that corresponded to the void fraction of the column (proteins >100 kDa) which also contained the majority of detectable PrPSc. Western blot detection of G100 fractionated PrPSc DRM-PK-PTA-G100 showed a major band in the void fraction #5 (B). A second PTA protein precipitation following G100 fractionation recovered detectable PrPSc in the void fractions (C). Evaluation of the purified PrPSc DRMPK-PTA-G100-PTA material by silver and Coomassie stain (D; left and middle part respectively) showed detectable PrPSc protein at the expected molecular weight that corresponded to PrPSc detection by Western blot (D; right part).
Figure 5 Increasing specific infectivity with PrPSc purification. Days of survival were determined by hamster bioassay for purified Scrapie brain homogenate preparations (A). Transmissible disease was observed following intracerebral inoculation of 1% PrPSc (25 µg), PrPSc DRM (1.35 µg) and PrPSc PK-treated PTA precipitated material fractionated by size exclusion on Sephadex G100 (PrPSc DRM-PK-PTA-G100; 0.8 µg). N = sample size. Comparison of ID50 and specific infectivity following intracerebral inoculation of 1% crude brain homogenate (brain), DRM and purified prion (DRM-PK-PTA-G100) by incubation time assay (B). Isolation of PrPSc in DRMs from lipid rafts results in >20-fold and purified PrPSc DRM-PK-PTA-G100 >40-fold, increase in specific infectivity relative to crude brain.
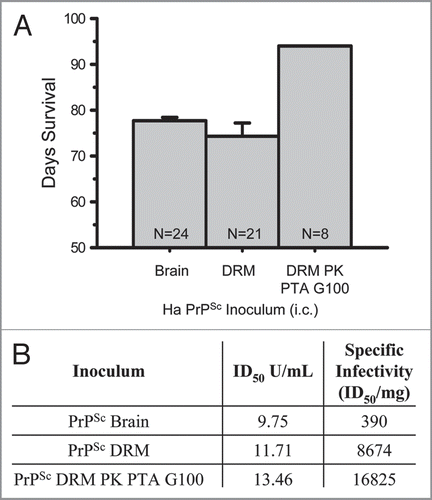
Figure 6 Schematic diagram of prion purification and immunization strategy. An outline of steps involved in prion purification from brain homogenate to immunization of Prnp0/0/Balbc/J and wild-type Balbc/J mice with purified PrPSc (PrPSc DRM-PK-PTA-G100-PTA).
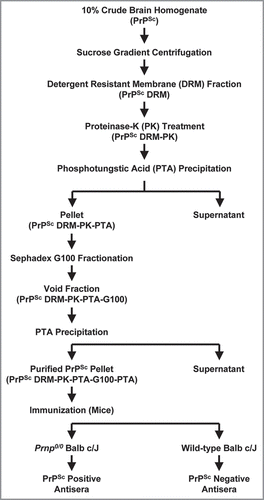
Figure 7 Detection of SH aPrP (90-231) by ELISA with antisera from prion immunized Prnp0/0/Balbc/J, but not wt Balbc/J mice. Antisera from Prnp0/0/Balbc/J mice detected SH aPrP (90-321) at dilutions >1:30 K by direct binding ELISA; whereas antisera from immunized wt Balbc/J mice failed to detect SH aPrP (90-231) at any dilution (A). Antisera from prion immunized Prnp0/0/Balbc/J, but not wt mice, detected SH aPrP (90-231), Syrian hamster brain PrPC (Ha PrPSc DRM; no PK) and PrPSc (Ha PrPSc DRM; +PK) by Western blot (B). Detection of PK-resistant PrPSc was observed with antisera diluted >25 K from two representative Prnp0/0/Balbc/J (#1–2) and wt Balbc/J (#3–4) mice immunized with purified prion antigen. RLU = relative light units.
Figure 8 Antisera from Prnp0/0/Balbc/J mice immunized with purified PrPSc binds Syrian hamster prion proteins. Western blot comparing binding of antisera from a prion imunized Prnp0/0/Balbc/J to normal and infectious hamster brain homogenate (30 µg/lane), recSH aPrP(90-231); 100 ng/lane) and normal and infectious hamster brain DRM (10 µg/lane) preparations (top part). Antisera detected PK-sensitive PrPC, recSH aPrP(90-231) and PK-insensitive PrPSc in brain homogenates and DRM preparations; no other protein bands were observed. A comparative western blot showed similar binding of the monoclonal anti-prion antibody IPC1 to brain homogenate, recombinant PrP and DRM preparations (bottom part) as antisera from Prnp0/0/Balbc/J mice. PK = proteinase-K treatment (+). Protein normalization by BCA.
Figure 9 Detection of endogenous prion proteins in normal (PrPC) and diseased (PrPSc) hamster brain DRM fractions by ELISA. Antisera from a prion immunized Prnp0/0/Balbc/J mouse (1:10K) bound DRM fraction from both PrPC and PrPSc in the absence of PK (−); black bars. Detection of PK-resistant PrP27-30 was observed in PrPSc, but not PrPC, DRM fractions following PK-treatment (+); open bars. No binding to any DRM fraction was observed with antisera from non-immunized Prnp0/0/Balbc/J mice. ELISA performed in triplicate and DRM protein normalized to 2 µg/well by BCA. RLU = relative light units. PK = proteinase-K.
Acknowledgements
This work was supported and administered under the USDA-ARS National Program in Animal Health (CRIS #5325-32000-008-00D), by a USDA Cooperative Agreement (58-5325-3-246), by grants from the National Institutes of Health (AG02132, AG10770 and AG021601) and by a gift from the G. Harold and Leila Y. Mathers Charitable Foundation.
References
- Prusiner SB. Prions. Proc Natl Acad Sci USA 1998; 23:13363 - 13363
- Bolton DC, McKinley MP, Prusiner SB. Identification of a protein that purifies with the scrapie prion. Science 1982; 4579:1309 - 1311
- Brandner S, Raeber A, Sailer A, Blattler T, Fischer M, Weissmann C, et al. Normal host prion protein (PrPC) is required for scrapie spread within the central nervous system. Proc Natl Acad Sci USA 1996; 23:13148 - 13151
- Prusiner SB, Scott M, Foster D, Pan KM, Groth D, Mirenda C, et al. Transgenetic studies implicate interactions between homologous PrP isoforms in scrapie prion replication. Cell 1990; 4:673 - 686
- MacGregor I. Prion protein and developments in its detection. Transfus Med 2001; 1:3 - 14
- McKinley MP, Bolton DC, Prusiner SB. A proteaseresistant protein is a structural component of the scrapie prion. Cell 1983; 1:57 - 62
- Meyer RK, McKinley MP, Bowman KA, Braunfeld MB, Barry RA, Prusiner SB. Separation and properties of cellular and scrapie prion proteins. Proc Natl Acad Sci USA 1986; 8:2310 - 2314
- Hadlow WJ, Kennedy RC, Race RE. Natural infection of Suffolk sheep with scrapie virus. J Infect Dis 1982; 5:657 - 664
- Hadlow WJ, Kennedy RC, Race RE, Eklund CM. Virologic and neurohistologic findings in dairy goats affected with natural scrapie. Vet Pathol 1980; 2:187 - 199
- Naslavsky N, Stein R, Yanai A, Friedlander G, Taraboulos A. Characterization of detergent-insoluble complexes containing the cellular prion protein and its scrapie isoform. J Biol Chem 1997; 10:6324 - 6331
- Kubler E, Oesch B, Raeber AJ. Diagnosis of prion diseases. Br Med Bull 2003; 267 - 279
- Tatzelt J, Groth DF, Torchia M, Prusiner SB, DeArmond SJ. Kinetics of prion protein accumulation in the CNS of mice with experimental scrapie. J Neuropathol Exp Neurol 1999; 12:1244 - 1249
- Pastrana MA, Sajnani G, Onisko B, Castilla J, Morales R, Soto C, et al. Isolation and characterization of a proteinase K-sensitive PrPSc fraction. Biochemistry 2006; 51:15710 - 15717
- Prusiner SB, Groth D, Serban A, Koehler R, Foster D, Torchia M, et al. Ablation of the prion protein (PrP) gene in mice prevents scrapie and facilitates production of anti-PrP antibodies. Proc Natl Acad Sci USA 1993; 22:10608 - 10612
- Williamson RA, Peretz D, Smorodinsky N, Bastidas R, Serban H, Mehlhorn I, et al. Circumventing tolerance to generate autologous monoclonal antibodies to the prion protein. Proc Natl Acad Sci USA 1996; 14:7279 - 7282
- Tsukamoto T, Diringer H, Ludwig H. Absence of autoantibodies against neurofilament proteins in the sera of scrapie infected mice. Tohoku J Exp Med 1985; 4:483 - 484
- Clarke MC, Haig DA. Attempts to demonstrate neutralising antibodies in the sera of scrapie-affected animals. Vet Rec 1966; 19:647 - 649
- Sargiacomo M, Scherer PE, Tang Z, Kubler E, Song KS, Sanders MC, et al. Oligomeric structure of caveolin: implications for caveolae membrane organization. Proc Natl Acad Sci USA 1995; 20:9407 - 9411
- Aguzzi A, Heikenwalder M, Miele G. Progress and problems in the biology, diagnostics and therapeutics of prion diseases. J Clin Invest 2004; 2:153 - 160
- Van EB, Boons J, De LE, Lubke U, Cras P. Molecular diagnostic tools in Creutzfeldt-Jakob disease and other prion disorders. Expert Rev Mol Diagn 2004; 3:351 - 359
- Hill AF, Collinge J. Subclinical prion infection. Trends Microbiol 2003; 12:578 - 584
- Prusiner SB, Scott MR, DeArmond SJ, Cohen FE. Prion protein biology. Cell 1998; 3:337 - 348
- Parveen I, Moorby J, Allison G, Jackman R. The use of non-prion biomarkers for the diagnosis of Transmissible Spongiform Encephalopathies in the live animal. Vet Res 2005; 5:665 - 683
- DeArmond SJ, Prusiner SB. Perspectives on prion biology, prion disease pathogenesis and pharmacologic approaches to treatment. Clin Lab Med 2003; 1:1 - 41
- Cohen FE, Prusiner SB. Pathologic conformations of prion proteins. Annu Rev Biochem 1998; 793 - 819
- Prusiner SB, Bolton DC, Groth DF, Bowman KA, Cochran SP, McKinley MP. Further purification and characterization of scrapie prions. Biochemistry 1982; 26:6942 - 6950
- Gabizon R, McKinley MP, Groth D, Prusiner SB. Immunoaffinity purification and neutralization of scrapie prion infectivity. Proc Natl Acad Sci USA 1988; 18:6617 - 6621
- Gregoire S, Logre C, Metharom P, Loing E, Chomilier J, Rosset MB, et al. Identification of two immunogenic domains of the prion protein-PrP-which activate class II-restricted T cells and elicit antibody responses against the native molecule. J Leukoc Biol 2004; 1:125 - 134
- Vey M, Pilkuhn S, Wille H, Nixon R, DeArmond SJ, Smart EJ, et al. Subcellular colocalization of the cellular and scrapie prion proteins in caveolae-like membranous domains. Proc Natl Acad Sci USA 1996; 25:14945 - 14949
- Stahl N, Borchelt DR, Hsiao K, Prusiner SB. Scrapie prion protein contains a phosphatidylinositol glycolipid. Cell 1987; 2:229 - 240
- Kaneko K, Vey M, Scott M, Pilkuhn S, Cohen FE, Prusiner SB. COOH-terminal sequence of the cellular prion protein directs subcellular trafficking and controls conversion into the scrapie isoform. Proc Natl Acad Sci USA 1997; 6:2333 - 2338
- Taraboulos A, Scott M, Semenov A, Avrahami D, Laszlo L, Prusiner SB. Cholesterol depletion and modification of COOH-terminal targeting sequence of the prion protein inhibit formation of the scrapie isoform. J Cell Biol 1995; 1:121 - 132
- Chesebro B, Trifilo M, Race R, Meade-White K, Teng C, LaCasse R, et al. Anchorless prion protein results in infectious amyloid disease without clinical scrapie. Science 2005; 5727:1435 - 1439
- Caughey B, Race RE, Ernst D, Buchmeier MJ, Chesebro B. Prion protein biosynthesis in scrapieinfected and uninfected neuroblastoma cells. J Virol 1989; 1:175 - 181
- Safar J, Wille H, Itri V, Groth D, Serban H, Torchia M, et al. Eight prion strains have PrPSc molecules with different conformations. Nat Med 1998; 10:1157 - 1165
- Wadsworth JD, Joiner S, Hill AF, Campbell TA, Desbruslais M, Luthert PJ, et al. Tissue distribution of protease resistant prion protein in variant Creutzfeldt-Jakob disease using a highly sensitive immunoblotting assay. Lancet 2001; 9277:171 - 180
- Bueler H, Aguzzi A, Sailer A, Greiner RA, Autenried P, Aguet M, et al. Mice devoid of PrP are resistant to scrapie. Cell 1993; 7:1339 - 1347
- Donne DG, Viles JH, Groth D, Mehlhorn I, James TL, Cohen FE, et al. Structure of the recombinant full-length hamster prion protein PrP(29-231): the N terminus is highly flexible. Proc Natl Acad Sci USA 1997; 25:13452 - 13457