Abstract
Prion proteins misfold and aggregate into multiple infectious strain variants that possess unique abilities to overcome prion species barriers, yet the structural basis for the species-specific infectivities of prion strains is poorly understood. Therefore, we have investigated the site-specific structural properties of a promiscuous chimeric form of the yeast prion Sup35 from Saccharomyces cerevisiae and Candida albicans. The Sup35 chimera forms two strain variants, each of which selectively infect one species but not the other. Importantly, the N-terminal and middle domains of the Sup35 chimera (collectively referred to as Sup35NM) contain two prion recognition elements (one from each species) that regulate the nucleation of each strain. Mutations in either prion recognition element significantly bias nucleation of one strain conformation relative to the other. Herein, we have investigated the folding of each prion recognition element for the serine-to-arginine mutant at residue 17 of Sup35NM chimera known to promote nucleation of C. albicans strain conformation. Using cysteine-specific labeling analysis, we find that residues in the C. albicans prion recognition element are solvent-shielded, while those outside the recognition sequence (including most of those in the S. cerevisiae recognition element) are solvent-exposed. Moreover, we find that proline mutations in the C. albicans recognition sequence disrupt the prion templating activity of this strain conformation. Our structural findings reveal that differential folding of complementary and non-complementary prion recognition elements within the prion amyloid core of the Sup35NM chimera is the structural basis for its species-specific templating activity.
Introduction
Several neurodegenerative disorders (e.g., Alzheimer and prion diseases) are related by improper folding and aggregation of specific proteins into similar types of β-sheet rich, oligomeric structures.Citation1,Citation2 However, Prion diseases are unique among conformational disorders since the mammalian prion protein PrP misfolds into infectious β-sheet rich, aggregated conformers that are linked to several fatal infectious diseases in mammals (transmissible spongiform encephalopathies).Citation2,Citation3 Many prions in yeast and other fungi have also been identified,Citation4–Citation16 the most-well studied of which is Sup35.Citation5–Citation8 Sup35 is a yeast translation-termination factor whose switch to its aggregated prion conformation reduces its activity, leading to an increase in read-through of stop codons and multiple phenotypic changes (e.g., change in colony color on adenine-deficient media).Citation5–Citation8,Citation17–Citation19 Sup35′s N-terminal (N) domain is highly amyloidogenic, and essential for prion formation and propagation.Citation5–Citation8 This domain is rich in glutamine and asparagine residues, and contains 5.5 imperfect oligopeptide repeats ([P/Q]QGGYQ[Q/S]YN) reminiscent of the mammalian PrP's oligopeptide repeats (P[H/Q]GGGWGQ).Citation20–Citation23 The middle (M) domain is rich in charged residues and acts to solubilize the N domain.Citation5–Citation8,Citation10 Sup35's C-terminal domain encodes its translation termination function.Citation5–Citation8,Citation10 Together, the N and M domains (NM) can form infectious prion conformers with or without the C-terminal domain of Sup35.Citation24,Citation25
Both PrP and Sup35 display two of the most important and puzzling aspects of prion biology. First, both proteins adopt not just one aggregated prion conformation,Citation26 but rather several related yet structurally distinct strain conformations.Citation19,Citation24,Citation27–Citation37 Each strain conformation self-perpetuates and confers a distinct biological phenotype.Citation24,Citation27–Citation31,Citation33,Citation38,Citation39 Second, the transmission of the prion state between proteins of different species is limited by a species barrier that can be occasionally crossed.Citation40–Citation55 Interestingly, the ability to establish and overcome species barriers in both yeast and mammals appears to be linked to the ability of prions to form distinct strains,Citation5,Citation37,Citation40,Citation44,Citation47,Citation49,Citation54–Citation63 yet the structural mechanisms linking prion strains and species barriers are poorly understood. In yeast, transmission of Sup35 prions from S. cerevisiae (Sc) to C. albicans (Ca) and vice versa is limited by a species barrier.Citation43,Citation46,Citation56,Citation60 However, a promiscuous chimeric Sup35 prion consisting of domains of Sc and Ca Sup35 (Fig. S1) can form two strain conformations with unique specificities to overcome this species barrier.Citation37,Citation43,Citation56,Citation60
Towards understanding how prion amyloid structure governs the selective infection of one species relative to another, we previously asked whether the nucleation of each strain conformation of the NM portion of the Sup35 chimera (herein referred to as the Sup35NM chimera) could be localized to one or more small sequence segments within the chimeric prion protein.Citation64 Using peptide microarrays that displayed hundreds of overlapping 20mer peptides from Sc and Ca Sup35, we identified two prion recognition sequences that regulated the nucleation of each strain conformation. We found that a mutation (S17R) in the S. cereviase prion recognition element which favored nucleation of a prion strain conformation specific for infecting C. albicansCitation43,Citation56 led to a highly specific interaction between the Sup35NM chimera and Ca peptides (residues 59–86).Citation64 In contrast, we found that mutations in the C. albicans recognition element (G70A, G71A, G80A, G81A) which promoted nucleation of a prion strain specific for infecting S. cerevisiaeCitation43,Citation56 led to specific association between the Sup35NM chimera and Sc peptides (residues 9–39).Citation64
Our discovery of two prion recognition elements within the Sup35NM chimera led us and othersCitation37 to hypothesize that the structural basis of the species-specific infectivities of its two chimeric prion strains is selective inclusion of one recognition element within the amyloid core, while the other one is excluded from the core. Indeed, recent amide exchange and proteolysis experiments for the wild-type Sup35NM chimera revealed that the S. cerevisiae strain conformation contains the Sc recognition sequence within its amyloid core, while the Ca recognition element is excluded from the amyloid core.Citation37 However, the C. albicans strain conformation was found not only to contain the Ca recognition sequence within its amyloid core, but the Sc prion recognition element was also partially solvent protected. The partial protection of the Sc recognition element suggests that it may be aggregated within the C. albicans strain conformation of the Sup35NM chimera. Since weCitation64 and othersCitation56,Citation65 had previously found that the S17R mutation in the Sc recognition element inhibited nucleation of the S. cerevisiae strain conformation of the Sup35NM chimera, we hypothesized that the S17R mutation would prevent aggregation of the Sc recognition element in the C. albicans strain conformation. Herein, we report our site-specific structural analysis of each prion recognition element within the C. albicans strain conformation of the S17R Sup35NM chimera.
Results and Discussion
We first sought to define the solvent-shielded residues within the folded prion core relative to those excluded from the core. As a site-specific approach for identifying residues within the amyloid core, we synthesized a panel of single cysteine variants of the S17R Sup35NM chimera that could be used to report the extent of solvent accessibility via cysteine-specific labeling after folding each mutant into prion amyloids. We generated 24 single cysteine substitution mutants distributed throughout the prion (N) and non-prion (M) domains (Fig. S1). Each purified Sup35NM chimera mutant was templated specifically into prion amyloids via seeding with CaNM fibrils (37°C). All mutants formed SDS-resistant amyloids that were indistinguishable via SDS-PAGE analysis (Fig. S2 and data not shown). We evaluated the templating activity of each cysteine mutant for specifically converting monomeric CaNM into prion fibrils relative to monomeric ScNM to evaluate if the cysteine mutations alter the biochemical activity of the chimeric prion amyloids. Importantly, all mutants display similar, significant seeding activity for CaNM monomers and negligible seeding activity for ScNM monomers ( and S3).
Once each cysteine mutant of the S17R Sup35NM chimera was assembled into the C. albicans strain conformation, we labeled the amyloids with a cysteine-specific dye (Lucifer yellow iodoacetamide) to identify solvent-shielded residues that are poorly reactive due to inclusion within the amyloid core.Citation39,Citation66 We also labeled monomers of each mutant to evaluate the fibril-to-monomer labeling ratio as a measure of the extent of solvent accessibility of each residue. Importantly, we first confirmed that Lucifer yellow fails to label the original S17R Sup35NM chimera that lacks cysteine (<5%; data not shown). In , we report the extent of labeling of each cysteine mutant in the C. albicans strain conformation of the S17R Sup35NM chimera. We find that residues within and near to the Ca recognition element (residues 59–86) are the most protected (<50% solvent exposed except for Q68), while those residues that flank this recognition element (including most of those in the Sc recognition element) are >50% solvent exposed (). Notably, we find that residue Q38 in the Sc recognition element is protected (∼20% solvent exposed), suggesting that the edge of the Sc recognition element folds within the amyloid core along with the Ca recognition element. Moreover, we find that other residues flanking the Ca recognition element (residues 88–142) also display a similar level of solvent exposure as the Sc recognition element. Finally, residues in the middle domain of the NM chimera (residues 168 and 213) are highly solvent accessible (>90% solvent exposed), consistent with the highly charged nature of this domain.Citation67,Citation68
We next evaluated the impact of the S17R mutation on the folding of the Sc prion recognition element within the C. albicans chimeric strain conformation by analyzing the wild-type Sup35NM chimera that lacks this mutation. We expected that the lack of the S17R mutation would decrease the solvent exposure of the Sc recognition element. Therefore, we repeated the labeling analysis for residues within the S. cerevisiae recognition element of the wild-type Sup35NM chimera in the C. albicans conformation. As expected, we find that the residues within the Sc recognition element are less solvent exposed for the wild-type Sup35NM chimera than for the S17R mutant ().
Next, we investigated the functional significance of the solvent-shielded residues within the C. albicans strain conformation of the S17R Sup35NM chimera using proline substitution mutagenesis. We sought to identify residues in the Sup35NM chimera involved in β-sheets (or other related folds) that are incompatible with proline residues.Citation38,Citation69 Thus, we evaluated the templating activity of the C. albicans strain conformation for converting single proline mutants of the Sup35NM chimera from monomers into fibrils. We hypothesized that residues normally folded within the amyloid core will be inefficiently folded into the core when mutated to proline.Citation38,Citation69,Citation70 Therefore, we synthesized a panel of 22 single proline substitution mutations in the N and M domains of the prion chimera, and evaluated the rate at which each mutant is templated into the prion state by the C. albicans strain conformation of the S17R Sup35NM chimera ().
We find that proline mutations within or near the Ca recognition element (residues 38–99) are most disruptive to the seeding activity of the S17R Sup35NM chimera (>20% defect; ). In contrast, proline mutations flanking the Ca recognition element (including most of those in the Sc recognition element) show little defect in templating activity, in agreement with the cysteine labeling experiments (). Mutating residue Q38 (at the edge of the Sc recognition element) to proline is also disruptive (∼25% seeding defect), consistent with the fact that it is typically folded within the amyloid core of this strain conformation. Finally, we find that some proline mutations, especially residues in the Sc recognition element (residues 10, 12 and 33), accelerate the templating activity of fibrils of the S17R Sup35NM chimera. This finding suggests that the wild-type Sc recognition element participates in interactions that lower the templating activity of the C. albicans strain conformation of the Sup35NM chimera, and these interactions are attenuated via proline mutations in the Sc recognition sequence. Overall, the relative impact of proline mutations on prion templating activity () is in excellent agreement with the cysteine labeling analysis (). These results suggest that residues in the Ca recognition element that are folded within the amyloid core directly mediate the species-specific templating activity of the C. albicans strain conformation.
A key outcome of our studies is site-specific structural details regarding the differential folding of the Sc and Ca prion recognition elements within one strain conformation of the promiscuous Sup35 prion chimera. There are several plausible structural models for how each recognition sequence could be differentially folded in the two strain conformations of the Sup35 chimera to yield unique capacities to overcome the S. cerevisiae/C. albicans species barrier. It is logical that the Ca recognition sequence is folded within the amyloid core of the C. albicans strain conformation (as reported previously in ref. Citation37), yet it is less clear how the highly amyloidogenic S. cerevisiae recognition sequence is inactive in this strain conformation. Prior to performing these experiments, we envisioned at least four potential structural mechanisms for the inactivity of the Sc recognition sequence in the C. albicans strain conformation of the Sup35 chimera: (1) all residues within the Sc recognition element are unfolded; (2) a subset of the Sc residues are unfolded while the rest are folded within the amyloid core; (3) Sc residues are in an alternative non-β-sheet folded conformation in which they can participate in intermolecular contacts; and (4) Sc residues are folded in an alternative β-sheet conformation in which they do not participate in intermolecular contacts necessary for templating activity. Our combined cysteine labeling and proline seeding results are most consistent with the Sc recognition element being largely unstructured except for the edge of this region (residue Q38) being folded within the amyloid core ().
Our results are complementary to those of Weissman and coworkers, who showed that the S. cerevisiae domain of the chimeric prion is partially solvent protected.Citation37 Importantly, this previous study demonstrated that the selective seeding of the C. albicans chimeric strain conformation does not require the S. cerevisiae domain, as it could be removed without altering seeding specificity or activity. Therefore, Weissman and coworkers conclude that the S. cerevisiae domain is in an aggregated, partially-protected conformation that is not competent for templating.Citation37 We also observe partial solvent protection of the Sc domain in the C. albicans strain conformation of the wild-type Sup35NM. Moreover, we find that the charged mutation S17R modestly increases the extent of solvent exposure of the Sc domain, especially within the Sc recognition element (). This finding is consistent with other reports that the S17R mutation prevents aggregation of Sc Sup35NMCitation56,Citation64 and Sup35,Citation65 and that this mutation strongly favors formation of the C. albicans strain conformation of the Sup35NM chimera.Citation56,Citation64,Citation65 Our results suggest that this charged mutation minimizes non-specific aggregation of the S. cerevisiae recognition element that otherwise occurs in the C. albicans strain conformation.Citation37 Our work reveals that prion amyloid structure is sensitive to single mutations within prion recognition elements.
Our findings also contribute to the broader understanding of how prion sequence, strain variation and species barriers are related. The amino acid sequence of a prion determines the spectrum of allowed strain conformations, and the ability of a donor prion to transmit its prion state to an acceptor prion is determined by whether their sequences encode compatible strain conformations.Citation5,Citation37,Citation41,Citation49,Citation51,Citation60,Citation70–Citation74 Our work (combined with that of Weissman and coworkersCitation37) provides a structural mechanism for defining the compatibility of prion strains formed by donor and acceptor prions. We find that inclusion of the Ca recognition element within the amyloid core of the Sup35NM chimera (donor) encodes its specific infectivity for CaNM (acceptor), and we posit that the Ca recognition element must be included within the CaNM amyloid core to form infectious C. albicans prions. Conversely, we find that partial exclusion of the Sc recognition sequence from the amyloid core of the prion chimera eliminated prion transmission to ScNM, which is consistent with previous findings that inclusion of the Sc recognition element within the amyloid core of ScNM is required for formation of infectious S. cerevisiae prions.Citation38,Citation39 Thus, we posit that the spectrum of allowed strain conformations is determined by proper folding of prion recognition elements within the amyloid core of donor prion strains that are complementary to the prion recognition sequences of the acceptor prion. This hypothesis would predict mutations in highly localized peptide segments (but not outside these segments) that impact the folding of prion recognition elements in the donor prion will govern the ability of such prions to cross species barriers. Indeed, we find that the S17R mutation impacts the folding of the non-complementary prion recognition element within the C. albicans strain conformation of the Sup35NM chimera. Moreover, additional point mutations in Sup35,Citation56,Citation65,Citation69,Citation75,Citation76 as well as in PrPCitation54,Citation58,Citation77 have been identified that both prevent and promote interspecies prion transmission. We expect that future structural studies aimed at investigating how mutations impact allowed strain conformations for closely related prions (such as PrPCitation78,Citation79 and Sup35Citation53,Citation71 variants) will reveal further insights into the role of the folding of small sequence elements within the prion amyloid core on the efficiency of interspecies prion transmission.
Materials and Methods
Mutagenesis and protein purification.
We used standard site-directed mutagenesis to generate single cysteine mutants of the Sup35NM chimera (both wild-type and S17R), as well as single proline mutants of the wild-type Sup35NM chimera. Each protein was purified via its C-terminal 7xHis tag, as described in references Citation64 and Citation68. Purified NM proteins were buffer exchanged into guanidinium hydrochloride (4 M, pH 7.4, 100 mM phosphate, 0.5 mM EDTA, 2 mM DTT) and stored at −80°C.
Fibril assembly, solubility and templating analysis.
We assembled fibrils of the Sup35NM chimera into the conformation specific for C. albicans by mixing monomeric wild-type or mutant chimeric protein of C. albicans NM (CaNM) fibrils overnight (20% w/v seed, PBS, 37°C, 1,200 rpm). Afterward, the chimeric fibrils were sedimented (110,000x g for 30 min at 4°C) and reconstituted in PBS. The SDS-resistance of fibrils was evaluated using a denaturing gel (NuPAGE 10% Bis-Tris gel) for prion samples before and after heating (100°C, 30 min). We evaluated the prion templating activity by following the rate at which chimeric prion fibrils (10% w/v) convert ScNM and CaNM monomers (2.5 µM) into Thioflavin T (ThT) positive conformers. We evaluated the ThT fluorescence (40 µM ThT) in 96-well plates using a SafireCitation2 plate reader (Tecan; λex = 450 nm, λem = 482 nm) to obtain initial seeding rates (∼10 min).
Cysteine accessibility measurements.
Fibrils of each cysteine chimera mutant (5 µM) were reduced with tris(2-carboxyethyl)phosphine (TCEP, 25 µM) and labeled with Lucifer yellow iodoacetamide (0.1 mM, PBS, pH 7.4) for 12 h at 25°C. Afterward, the labeled fibrils were centrifuged (110,000x g for 30 min at 4°C), dissociated (4 M GuHCl, 100°C, 5 min) and excess dye was removed (Zeba 96-well spin-desalting plates, Pierce Thermo Scientific). As a control, fibrils were also dissociated into monomers (4 M GuHCl, 100°C, 5 min) prior to labeling, and then monomers were labeled in the same manner as fibrils. The extent of labeling for both fibrils and monomers was evaluated using fluorescence (λex = 426 nm, λem = 531 nm) and protein (micro BCA assay kit, Pierce Thermo Scientific) assays.
Proline mutant seeding defect analysis.
Fibrils of the S17R Sup35NM chimera (10% w/v) were mixed with monomeric proline mutants (2.5 µM, PBS, pH 7.4), and their ThT fluorescence was monitored (12 h). The initial seeding rates of each proline mutant were compared to the wild-type Sup35NM chimera and reported as % defect as described previously in reference Citation38.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Figures and Tables
Figure 1 Templating specificity of the C. albicans prion strain conformation of the S17R Sup35NM chimera. The S17R Sup35NM chimera (Chim) and cysteine mutants thereof were assembled into prion amyloids and their templating specificity was analyzed using ThT fluorescence. The reported seeding activity of the chimeric Sup35NM fibrils is after subtraction of the contribution from the residual CaNM fibrils used to template the assembly of the Sup35NM chimera.
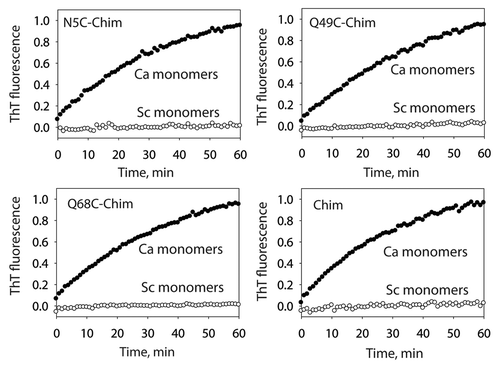
Figure 2 Solvent accessibility of cysteine residues in the C. albicans prion strain conformation of the Sup35NM chimera. (A) S17R Sup35NM chimera fibrils in the C. albicans strain conformation (as well as the corresponding monomers) were labeled using Lucifer yellow iodoacetamide. (B) Comparison of cysteine labeling of S17R and wild-type Sup35NM chimera fibrils in the S. cerevisiae domain. Each measurement is the average (±standard deviation) of at least three independent experiments.
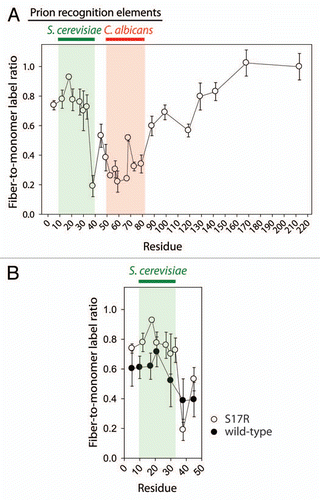
Figure 3 Impact of proline substitution mutants on the templating activity of the C. albicans strain conformation of the S17R Sup35NM chimera. S17R Sup35NM fibrils in the C. albicans strain conformation were evaluated for their activity to template proline substitution mutants of the Sup35NM chimera into prion fibrils. Each seeding rate (k) was calculated from the initial slope of the ThT measurements, and expressed as percent defect [100 ×(1 − kmut/kwt)], as described previously in reference Citation38.
![Figure 3 Impact of proline substitution mutants on the templating activity of the C. albicans strain conformation of the S17R Sup35NM chimera. S17R Sup35NM fibrils in the C. albicans strain conformation were evaluated for their activity to template proline substitution mutants of the Sup35NM chimera into prion fibrils. Each seeding rate (k) was calculated from the initial slope of the ThT measurements, and expressed as percent defect [100 ×(1 − kmut/kwt)], as described previously in reference Citation38.](/cms/asset/485a5ea8-f785-44a7-bdb5-f49ff44e2337/kprn_a_10916694_f0003.gif)
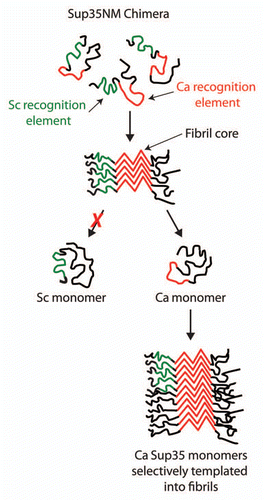
Figure 4 Selective folding of prion recognition elements within the Sup35NM chimera enciphers species-specific infectivity. The Sup35NM chimera contains two prion recognition sequences (green for the S. cerevisiae recognition element and red for C. albicans recognition element) that selectively nucleate each prion strain conformation. For the C. albicans strain conformation, we find that the Ca recognition sequence is folded within the prion amyloid core, while most of the Sc recognition sequence is excluded from the core. The resulting prion strain conformation is specific for infecting C. albicans Sup35 since its cognate recognition element is folded within the amyloid core and active for templating.
Additional material
Download Zip (1.5 MB)Acknowledgments
We thank Ali Reza Ladiwala, Joseph Perchiacca and Shantanu Sule for assistance editing the manuscript. This work was funded by the NIH (NS065333) and a Pew Scholars Award in Biomedical Sciences.
References
- Selkoe DJ. Alzheimer's disease. In the beginning. Nature 1991; 354:432 - 433
- Prusiner SB. Prions. Proc Natl Acad Sci USA 1998; 95:13363 - 13383
- Aguzzi A, Polymenidou M. Mammalian prion biology: one century of evolving concepts. Cell 2004; 116:313 - 327
- Wickner RB. [URE3] as an altered URE2 protein: evidence for a prion analog in Saccharomyces cerevisiae. Science 1994; 264:566 - 569
- Chien P, Weissman JS, DePace AH. Emerging principles of conformation-based prion inheritance. Annu Rev Biochem 2004; 73:617 - 656
- Shorter J, Lindquist S. Prions as adaptive conduits of memory and inheritance. Nat Rev Genet 2005; 6:435 - 450
- Tuite MF, Cox BS. Propagation of yeast prions. Nat Rev Mol Cell Biol 2003; 4:878 - 890
- Uptain SM, Lindquist S. Prions as protein-based genetic elements. Annu Rev Microbiol 2002; 56:703 - 741
- Coustou V, Deleu C, Saupe S, Begueret J. The protein product of the het-s heterokaryon incompatibility gene of the fungus Podospora anserina behaves as a prion analog. Proc Natl Acad Sci USA 1997; 94:9773 - 9778
- Wickner RB, Edskes HK, Shewmaker F, Nakayashiki T. Prions of fungi: inherited structures and biological roles. Nat Rev Microbiol 2007; 5:611 - 618
- Du Z, Park KW, Yu H, Fan Q, Li L. Newly identified prion linked to the chromatin-remodeling factor Swi1 in Saccharomyces cerevisiae. Nat Genet 2008; 40:460 - 465
- Brown JC, Lindquist S. A heritable switch in carbon source utilization driven by an unusual yeast prion. Genes Dev 2009; 23:2320 - 2332
- Nemecek J, Nakayashiki T, Wickner RB. A prion of yeast metacaspase homolog (Mca1p) detected by a genetic screen. Proc Natl Acad Sci USA 2009; 106:1892 - 1896
- Patel BK, Gavin-Smyth J, Liebman SW. The yeast global transcriptional co-repressor protein Cyc8 can propagate as a prion. Nat Cell Biol 2009; 11:344 - 349
- Alberti S, Halfmann R, King O, Kapila A, Lindquist S. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell 2009; 137:146 - 158
- Bradley ME, Edskes HK, Hong JY, Wickner RB, Liebman SW. Interactions among prions and prion “strains” in yeast. Proc Natl Acad Sci USA 2002; 99:16392 - 16399
- True HL, Berlin I, Lindquist SL. Epigenetic regulation of translation reveals hidden genetic variation to produce complex traits. Nature 2004; 431:184 - 187
- True HL, Lindquist SL. A yeast prion provides a mechanism for genetic variation and phenotypic diversity. Nature 2000; 407:477 - 483
- Eaglestone SS, Cox BS, Tuite MF. Translation termination efficiency can be regulated in Saccharomyces cerevisiae by environmental stress through a prion-mediated mechanism. EMBO J 1999; 18:1974 - 1981
- Liu JJ, Lindquist S. Oligopeptide-repeat expansions modulate ‘protein-only’ inheritance in yeast. Nature 1999; 400:573 - 576
- Parham SN, Resende CG, Tuite MF. Oligopeptide repeats in the yeast protein Sup35p stabilize intermolecular prion interactions. EMBO J 2001; 20:2111 - 2119
- Vital C, Gray F, Vital A, Parchi P, Capellari S, Petersen RB, et al. Prion encephalopathy with insertion of octapeptide repeats: the number of repeats determines the type of cerebellar deposits. Neuropathol Appl Neurobiol 1998; 24:125 - 130
- Ter-Avanesyan MD, Dagkesamanskaya AR, Kushnirov VV, Smirnov VN. The SUP35 omnipotent suppressor gene is involved in the maintenance of the non-Mendelian determinant [psi+] in the yeast Saccharomyces cerevisiae. Genetics 1994; 137:671 - 676
- Tanaka M, Chien P, Naber N, Cooke R, Weissman JS. Conformational variations in an infectious protein determine prion strain differences. Nature 2004; 428:323 - 328
- Krzewska J, Tanaka M, Burston SG, Melki R. Biochemical and functional analysis of the assembly of full-length Sup35p and its prion-forming domain. J Biol Chem 2007; 282:1679 - 1686
- Shorter J, Lindquist S. Destruction or potentiation of different prions catalyzed by similar Hsp104 remodeling activities. Mol Cell 2006; 23:425 - 438
- Bruce ME, McConnell I, Fraser H, Dickinson AG. The disease characteristics of different strains of scrapie in Sinc congenic mouse lines: implications for the nature of the agent and host control of pathogenesis. J Gen Virol 1991; 72:595 - 603
- Caughey B, Raymond GJ, Bessen RA. Strain-dependent differences in beta-sheet conformations of abnormal prion protein. J Biol Chem 1998; 273:32230 - 32235
- Derkatch IL, Chernoff YO, Kushnirov VV, Inge-Vechtomov SG, Liebman SW. Genesis and variability of [PSI] prion factors in Saccharomyces cerevisiae. Genetics 1996; 144:1375 - 1386
- King CY, Diaz-Avalos R. Protein-only transmission of three yeast prion strains. Nature 2004; 428:319 - 323
- Kocisko DA, Come JH, Priola SA, Chesebro B, Raymond GJ, Lansbury PT, Caughey B. Cell-free formation of protease-resistant prion protein. Nature 1994; 370:471 - 474
- Pattison IH, Jones KM. Modification of a strain of mouse-adapted scrapie by passage through rats. Res Vet Sci 1968; 9:408 - 410
- Safar J, Wille H, Itri V, Groth D, Serban H, Torchia M, et al. Eight prion strains have PrP(Sc) molecules with different conformations. Nat Med 1998; 4:1157 - 1165
- Somerville RA, Oberthur RC, Havekost U, MacDonald F, Taylor DM, Dickinson AG. Characterization of thermodynamic diversity between transmissible spongiform encephalopathy agent strains and its theoretical implications. J Biol Chem 2002; 277:11084 - 11089
- Chernoff YO, Derkach IL, Inge-Vechtomov SG. Multicopy SUP35 gene induces de-novo appearance of psi-like factors in the yeast Saccharomyces cerevisiae. Curr Genet 1993; 24:268 - 270
- Legname G, Nguyen HO, Baskakov IV, Cohen FE, Dearmond SJ, Prusiner SB. Strain-specified characteristics of mouse synthetic prions. Proc Natl Acad Sci USA 2005; 102:2168 - 2173
- Foo CK, Ohhashi Y, Kelly MJ, Tanaka M, Weissman JS. Radically different amyloid conformations dictate the seeding specificity of a chimeric sup35 prion. J Mol Biol 2011; 408:1 - 8
- Toyama BH, Kelly MJ, Gross JD, Weissman JS. The structural basis of yeast prion strain variants. Nature 2007; 449:233 - 237
- Krishnan R, Lindquist SL. Structural insights into a yeast prion illuminate nucleation and strain diversity. Nature 2005; 435:765 - 772
- Bruce KL, Chernoff YO. Sequence specificity and fidelity of prion transmission in yeast. Semin Cell Dev Biol 2011;
- Engel A, Shewmaker F, Edskes HK, Dyda F, Wickner RB. Amyloid of the Candida albicans Ure2p Prion Domain Is Infectious and Has an In-Register Parallel beta-Sheet Structure. Biochemistry 2011; 50:5971 - 5978
- Chernoff YO, Galkin AP, Lewitin E, Chernova TA, Newnam GP, Belenkiy SM. Evolutionary conservation of prion-forming abilities of the yeast Sup35 protein. Mol Microbiol 2000; 35:865 - 876
- Chien P, Weissman JS. Conformational diversity in a yeast prion dictates its seeding specificity. Nature 2001; 410:223 - 227
- Collinge J. Prion diseases of humans and animals: their causes and molecular basis. Annu Rev Neurosci 2001; 24:519 - 550
- Kushnirov VV, Kochneva-Pervukhova NV, Chechenova MB, Frolova NS, Ter-Avanesyan MD. Prion properties of the Sup35 protein of yeast Pichia methanolica. EMBO J 2000; 19:324 - 331
- Santoso A, Chien P, Osherovich LZ, Weissman JS. Molecular basis of a yeast prion species barrier. Cell 2000; 100:277 - 288
- Bruce M, Chree A, McConnell I, Foster J, Pearson G, Fraser H. Transmission of bovine spongiform encephalopathy and scrapie to mice: strain variation and the species barrier. Philos Trans R Soc Lond B Biol Sci 1994; 343:405 - 411
- Nakayashiki T, Ebihara K, Bannai H, Nakamura Y. Yeast [PSI+] “prions” that are crosstransmissible and susceptible beyond a species barrier through a quasi-prion state. Mol Cell 2001; 7:1121 - 1130
- Prusiner SB, Scott M, Foster D, Pan KM, Groth D, Mirenda C, et al. Transgenetic studies implicate interactions between homologous PrP isoforms in scrapie prion replication. Cell 1990; 63:673 - 686
- Resende C, Parham SN, Tinsley C, Ferreira P, Duarte JA, Tuite MF. The Candida albicans Sup35p protein (CaSup35p): function, prion-like behaviour and an associated polyglutamine length polymorphism. Microbiology 2002; 148:1049 - 1060
- Scott M, Foster D, Mirenda C, Serban D, Coufal F, Walchli M, et al. Transgenic mice expressing hamster prion protein produce species-specific scrapie infectivity and amyloid plaques. Cell 1989; 59:847 - 857
- Supattapone S, Bosque P, Muramoto T, Wille H, Aagaard C, Peretz D, et al. Prion protein of 106 residues creates an artificial transmission barrier for prion replication in transgenic mice. Cell 1999; 96:869 - 878
- Chen B, Newnam GP, Chernoff YO. Prion species barrier between the closely related yeast proteins is detected despite coaggregation. Proc Natl Acad Sci USA 2007; 104:2791 - 2796
- Collinge J, Palmer MS, Sidle KC, Hill AF, Gowland I, Meads J, et al. Unaltered susceptibility to BSE in transgenic mice expressing human prion protein. Nature 1995; 378:779 - 783
- Vishveshwara N, Liebman SW. Heterologous cross-seeding mimics cross-species prion conversion in a yeast model. BMC Biol 2009; 7:26
- Chien P, DePace AH, Collins SR, Weissman JS. Generation of prion transmission barriers by mutational control of amyloid conformations. Nature 2003; 424:948 - 951
- Collinge J, Sidle KC, Meads J, Ironside J, Hill AF. Molecular analysis of prion strain variation and the aetiology of ‘new variant’ CJD. Nature 1996; 383:685 - 690
- Hill AF, Desbruslais M, Joiner S, Sidle KCL, Gowland I, Collinge J, et al. The same prion strain causes vCJD and BSE. Nature 1997; 389:448 - 450
- Peretz D, Williamson RA, Legname G, Matsunaga Y, Vergara J, Burton DR, et al. A change in the conformation of prions accompanies the emergence of a new prion strain. Neuron 2002; 34:921 - 932
- Tanaka M, Chien P, Yonekura K, Weissman JS. Mechanism of cross-species prion transmission: an infectious conformation compatible with two highly divergent yeast prion proteins. Cell 2005; 121:49 - 62
- Will RG, Ironside JW, Hornlimann B, Zeidler M. Creutzfeldt-Jakob disease. Lancet 1996; 347:65 - 66
- Dickinson AG, Meikle VM. A comparison of some biological characteristics of the mouse-passaged scrapie agents, 22A and ME7. Genet Res 1969; 13:213 - 225
- Tessier PM, Lindquist S. Unraveling infectious structures, strain variants and species barriers for the yeast prion [PSI+]. Nat Struct Mol Biol 2009; 16:598 - 605
- Tessier PM, Lindquist S. Prion recognition elements govern nucleation, strain specificity and species barriers. Nature 2007; 447:556 - 561
- DePace AH, Santoso A, Hillner P, Weissman JS. A critical role for amino-terminal glutamine/asparagine repeats in the formation and propagation of a yeast prion. Cell 1998; 93:1241 - 1252
- Shivaprasad S, Wetzel R. Scanning cysteine mutagenesis analysis of Abeta-(1–40) amyloid fibrils. J Biol Chem 2006; 281:993 - 1000
- Liu JJ, Sondheimer N, Lindquist SL. Changes in the middle region of Sup35 profoundly alter the nature of epigenetic inheritance for the yeast prion [PSI+]. Proc Natl Acad Sci USA 2002; 99:16446 - 16453
- Glover JR, Kowal AS, Schirmer EC, Patino MM, Liu JJ, Lindquist S. Self-seeded fibers formed by Sup35, the protein determinant of [PSI+], a heritable prion-like factor of S. cerevisiae. Cell 1997; 89:811 - 819
- Chang HY, Lin JY, Lee HC, Wang HL, King CY. Strain-specific sequences required for yeast [PSI+] prion propagation. Proc Natl Acad Sci USA 2008; 105:13345 - 13350
- Ohhashi Y, Ito K, Toyama BH, Weissman JS, Tanaka M. Differences in prion strain conformations result from non-native interactions in a nucleus. Nat Chem Biol 2010; 6:225 - 230
- Chen B, Bruce KL, Newnam GP, Gyoneva S, Romanyuk AV, Chernoff YO. Genetic and epigenetic control of the efficiency and fidelity of cross-species prion transmission. Mol Microbiol 2010; 76:1483 - 1499
- Kocisko DA, Priola SA, Raymond GJ, Chesebro B, Lansbury PT Jr, Caughey B. Species specificity in the cell-free conversion of prion protein to protease-resistant forms: a model for the scrapie species barrier. Proc Natl Acad Sci USA 1995; 92:3923 - 3927
- Wickner RB, Edskes HK, Shewmaker F, Kryndushkin D, Nemecek J. Prion variants, species barriers, generation and propagation. J Biol 2009; 8:47
- Edskes HK, McCann LM, Hebert AM, Wickner RB. Prion variants and species barriers among Saccharomyces Ure2 proteins. Genetics 2009; 181:1159 - 1167
- King CY. Supporting the structural basis of prion strains: induction and identification of [PSI] variants. J Mol Biol 2001; 307:1247 - 1260
- Chernoff YO. Identity determinants of infectious proteins. Proc Natl Acad Sci USA 2008; 105:13191 - 13192
- Asante EA, Linehan JM, Desbruslais M, Joiner S, Gowland I, Wood AL, et al. BSE prions propagate as either variant CJD-like or sporadic CJD-like prion strains in transgenic mice expressing human prion protein. EMBO J 2002; 21:6358 - 6366
- Bartz JC, McKenzie DI, Bessen RA, Marsh RF, Aiken JM. Transmissible mink encephalopathy species barrier effect between ferret and mink: PrP gene and protein analysis. J Gen Virol 1994; 75:2947 - 2953
- Raymond GJ, Bossers A, Raymond LD, O'Rourke KI, McHolland LE, Bryant PK 3rd, et al. Evidence of a molecular barrier limiting susceptibility of humans, cattle and sheep to chronic wasting disease. EMBO J 2000; 19:4425 - 4430