Abstract
Phosphoinositides play important roles in eukaryotic cells, although they constitute a minor fraction of total cellular lipids. Specific kinases and phosphatases function on the regulation of phosphoinositide levels. Phosphatidylinositol 3-phosphate (PtdIns3P), a molecule of phosphoinositides regulates multiple aspects of plant growth and development. In this article, we introduce and discuss the kinases and phosphatases involved in PtdIns3P metabolism and their roles in pollen development and pollen tube growth in Arabidopsis.
Introduction
Phosphatidylinositols (PtdIns) are minor components in the eukaryotic cell membrane. The inositol ring of PtdIns can be phosphorylated at D3, D4, and D5 by specific phosphoinositide kinases, resulting in formation of phosphatidylinositol phosphate (PtdInsP), phosphatidylinositol bisphosphate (PtdInsP2) and phosphatidylinositol trisphosphate (PtdInsP3). These inositol phospholipids are called phosphoinositides (PIs). The lipid phosphatases remove the D3, D4, or D5 phosphates from these PIs to establish PI homeostatic levels in cell. Each PI isoform shows a specific pattern of intracellular distribution.
Fluorescence protein labeling has revealed that phosphatidylinositol 3-phosphate (PtdIns3P) mainly distributes in late endosomes, multivesicular bodies, and prevacuolar membranes.Citation1,Citation2 PIs themselves or serving as precursors for second messengers regulate plant growth and development, such as pollen development, pollen hydration and germination, pollen tube growth, root hair elongation, and stomatal movement modulation.Citation3-Citation12 Here, we focus on reviewing the functions of kinases and phosphatases in the PtdIns3P metabolism involved in pollen development, germination, and pollen tube growth in Arabidopsis.
The conversion between PtdIns and PtdIns3P
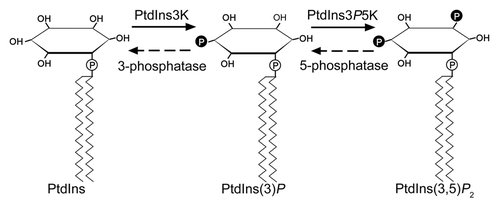
PtdIns can be phosphorylated to PtdIns3P by class III PtdIns 3-kinases (PtdIns3K) (). VPS34 is the first identified class III PtdIns3K in yeast with lipid kinase activity.Citation13 Two kinds of PtdIns3K complexes were identified in yeast and mammals. Complex I is composed of vacuolar protein sorting-associated protein (VPS) 34, VPS15, VPS30/autophagy-related protein (ATG) 6/coiled-coil myosin-like BCL2-interacting protein (BECLIN) 1 and ATG14, and regulates vesicle nucleation in autophagy. Complex II consists of VPS34, VPS15, VPS30/ATG6/BECLIN 1 and VPS38/UV irradiation resistance-associated protein (UVRAG), and functions on hydrolase sorting through the vacuolar protein sorting (VPS) pathway.Citation14,Citation15 By interacting directly with VPS34, VPS15 regulates the membrane targeting and activity of VPS34.Citation16-Citation18 VPS30/ATG6 was identified by searching AuTophaGy-related mutants in yeast. Mammalian Beclin1 is a homolog of yeast ATG6.Citation19 Although Vps30/Atg6/Beclin1 is found to be involved in autophagy and vesicular trafficking, its roles in PtdIns3K complexes are little known.Citation20,Citation21 ATG14 together with ATG6 mediates the localization of complex I and other ATG proteins on phagophore assembly site (PAS).Citation22 VPS38 serves as a bridge between VPS34-VPS15 and VPS30, and localizes complex II to endosome.Citation22,Citation23
Figure 1. Metabolism of Phosphatidylinositol 3-phosphate. PtdIns3K, PtdIns 3-kinase; PtdIns3P5K, PtdIns3P 5-kinase.
AtVPS34, AtVPS15 and AtVPS30 in Arabidopsis are the homologs of yeast VPS34, VPS15 and VPS30 (). The knockout of AtVPS34 in Arabidopsis plants leads to pollen failure in undergoing normal fission after the first mitotic division. Although the morphology of the atvps34 mutant is normal, its pollens display defects in vacuolar morphology and cannot germinate.Citation8 Either atvps15 or atvps30 mutant also shows abnormal pollen germination. The defect of atvps15 pollen germination can be restored partially after applification of PtdIns3P in vitro.Citation11,Citation24-Citation26 Therefore, AtVPS34, AtVPS15 and AtVPS30 might be the components of PtdIns3K complex in plant cells as in yeast, and function in PtdIns3P generation and pollen tube growth. PtdIns3P is mainly localized in endosomes and vacuolar membrane in plant cells.Citation2,Citation7 Although both atvps15 and atvps30 mutants displays abnormal germination of pollen, no vacuolar phenotype in atvps30 pollen was observed.Citation24-Citation26 It is likely that AtVPS30 in the PtdIns3K complex are not involved in PtdIns3P metabolism. Alternatively, AtVPS15 and AtVPS34, but not AtVPS30, may regulate vacuolar development in pollen development. It was reported that AtVPS30 is co-localized with ATATG8, a PAS marker protein in Arabidopsis.Citation24 AtVPS30 may be involved in vesicular trafficking by recruiting the PtdIns3K complex to the putative PAS in Arabidopsis plant cells, which is required for pollen germination and pollen tube growth. Three putative homologs of ATG14/UVRAG were identified in Arabidopsis (gene ID: AT2G32760, AT4G08540, AT1G77890; ) according to their putative protein containing the conserved ATG14 domain. These indicate that class III PtdIns3K complex is conserved in animals, yeast and plants. Two genes (ID: AT4G08540, AT1G77890) are predicated to encode the DNA-directed RNA polymerase. Another gene’s (ID: AT2G32760) function is unknown although its expression is detected during pollen development (bbc.botany.utoronto.ca/efp/cgi-bin/efpWeb.cgi). The components of class III PtdIns3K complex and their functions remain to be investigated.
Table 1. Enzymes and regulators for PI3P metabolism in Saccharomyces cerevisiae and Homo sapiens, and their homologs in Arabidopsis.
PtdIns3P 3-phosphatases can cleave the 3-phosphate from PtdIns3P and form PtdIns ().Citation27 The level of PtdIns3P decreases in yeast that expresses myotubularin-related protein (MTMR) 3, a human PtdIns3P 3-phosphatases belonging to the myotubularin family.Citation28 Myotubularins (MTMs) is a lipid phosphatase family with specificity for PtdIns3P and phosphatidylinositol 3,5-bisphosphate (PtdIns(3,5)P2) as substrates.Citation29 Yeast myotubularin-related protein (YMR) 1, the only myotubularin 3-phosphatase in yeast, corporately functions with synaptojanin-like phosphatase synaptojanin-like protein (SJL) 3 in regulating the localization and level of PtdIns3P.Citation30 In human, 14 members of myotubularin family PtdIns3P 3-phosphatases were identified, and their mutation led to human diseases.Citation31 ATMTM1 and ATMTM2 in Arabidopsis are myotubularin family homologs ().Citation32,Citation33 ATMTM1 dephosphorylates for PtdIns3P, however its phosphatase activity is higher with PtdIns(3,5)P2 as substrate than with PtdIns3P in vitro. Gene expression analysis reveals that the transcription of AtMTM1 increases significantly after drought stress, indicating its function might be involved in a drought responsive pathway.Citation32 ATMTM2 shows high expression level in pollen (www.bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi), but its roles in pollen development and pollen dehydration remains to be established.
PTEN/MMAC (a phosphatase and tensin homolog / mutated in multiple advanced cancers), dephosphorylating the D3 phosphate group of phosphatidylinositol 3,4,5-trisphosphate (PtdIns(3,4,5)P3), functions in autophagy of mammalian.Citation34,Citation35 In Arabidopsis genome, there are three putative PTEN homologs, AtPTEN1, AtPTEN2A and AtPTEN2B, and they show high expressions in pollen.Citation36,Citation37 The silencing of AtPTEN1 by RNA interference led to aborted pollen grains.Citation36 Additionally, the overexpression of either AtPTEN1 or AtPTEN2A induces male sterility.Citation12,Citation37 Although AtPTEN1 can dephosphorylate PtdIns(3,4,5)P3 in vitro, it still can bind PtdIns3P. The overexpression of AtPTEN1 caused the accumulation of autophagic bodies in tobacco pollen tube, which can be inhibited by exogenous PtdIns3P or by the expression of AtVPS34.Citation12 Thus, AtPTEN1 regulating PtdIns3P metabolism is involved in the autophagy in pollen tube.
The Conversion between PtdIns3P and PtdIns(3,5)P2
Class III PtdIns3P 5-kinases (PtdIns3P5K) catalyze the phosphorylation of PtdIns3P to PtdIns(3,5)P2 () that plays vital roles in endosomal trafficking.Citation38 FYVE (FAB1, YGL023, VPS27 and EEA1) domain-containing proteins, yeast formation of haploid and binucleate cells 1 (FAB1) and mammal FYVE finger-containing phosphoinositide kinase (PIKfyve), exhibit PtdIns3P5K activity.Citation39 Mutation of FAB1 reduces PtdIns(3,5)P2 level in yeast. The PtdIns3P5K activity of FAB1 is required for cargo-selective sorting into the vacuolar lumen in yeast. PIKfyve regulates the retrograde traffic from endosome to trans-Golgi-network.Citation40,Citation41 Arabidopsis genome contains two putative FAB1/PIKfyve homologs, AtFAB1A and AtFAB1B.Citation42 The pollen of the double mutant atfab1a atfab1b is lethal and shows severe defects in vacuolar organization.Citation43 The enhanced or reduced levels of AtFAB1A/B expression interfere the endomembrane homeostasis including endocytosis, vacuolar formation and acidification.Citation44 In yeast, hyperosmotic stress increases the level of PtdIns(3,5)P2 that plays roles in membrane trafficking in the endosomal/lysosomal system.Citation38 Endocytosis is also involved in pollen tube growth.Citation45 Therefore, PtdIns(3,5)P2 might play roles through similar mechanism in pollen tube growth with yeast hyperosmotic tolerance.
FAB1/PIKfyve forms a protein complex with factor-induced protein (FIG) 4/suppressor of actin (SAC) 3, vacuolar segregation protein (VAC) 7, ATG18 and VAC14 in yeast.Citation46,Citation47 Homologs of the subunits of the FAB1/PIKfyve complex except VAC7 were identified in Arabidopsis (). Although VAC7 is a FAB1 activator in PtdIns(3,5)P2-synthesizing complex, hyperosmotic shock still induces the increase in PtdIns(3,5)P2 levels in the absence of VAC7 in yeast.Citation48 Thus, VAC7 is not necessary in this complex for PtdIns(3,5)P2 synthesis and turnover. Whereas, FIG4 itself can function in both PtdIns(3,5)P2 synthesis and turnover in yeast. Function of the most Arabidopsis homologs of FAB1/PIKfyve complex components are not investigated, except that the FIG4 homolog (gene ID: At5G66020, ) was found to be involved in pollen development.Citation49
PtdIns(3,5)P2 5-phosphatase hydrolyzes the 5-phosphate of PtdIns(3,5)P2 to form PtdIns3P (). In yeast, the level of PtdIns(3,5)P2 significantly increases after hyperosmotic stress, and then, it decreases to the basal level after 30 min. It was found that yeast FIG4, a SAC phosphatase domain-containing protein, functions in this process.Citation50 In the cells lacking FIG4, PtdIns(3,5)P2 level shows a little decrease after hyperosmotic shock. Other proteins, SAC1, SJL1, SJL2 and SJL3, also show similarity with FIG4 in yeast.Citation51 Synaptojanin is the mammalian phosphoinositide 5-phosphatases, such as hSAC1, hSAC2, hSAC3, Synaptojanin 1 and Synaptojanin 2 in human. The loss of function of hSAC3, a homolog of FIG4 in human, reduces the level of PtdIns(3,5)P2 and leads to neuronal degeneration.Citation52,Citation53 Arabidopsis genome encodes nine putative SAC domain-containing proteins (AtSACs) that can be classified into three classes: AtSAC1–5 are similar to yeast FIG4, AtSAC6–8 are similar to yeast SAC1, and AtSAC9 contains unique domains ().Citation54 AtSAC1 exhibits phosphatase activity with PtdIns(3,5)P2 as the substrate. Mutation of AtSAC1 causes a decrease in the wall thickness, and results in a weak stem phenotype.Citation49 However, AtSAC7 (ROOT HAIR DEFECTIVE 4) and AtSAC9 all display a preference for phosphatidylinositol 4,5-bisphosphate (PtdIns(4,5)P2) as substrate in vitro.Citation55,Citation56 Mutation of both AtSAC7 and AtSAC9 lead to overaccumulation of PtdIns(4,5)P2, resulted in root hair-defect and constitutive expression of the stress-response genes. Five SAC-like genes (gene ID At1G22620, At3G51460, At3G59770, At3G14205, At5G20840 and At5G66020) show high expression level in Arabidopsis pollen ().Citation57 Only the function of AtSAC6 was found to be involved in β-aminobutyric acid-induced pollen sterility.Citation58
Conclusion and Perspective
Although some information has been accumulated on understanding for PtdIns3P metabolism in Arabidopsis, the functions of itself and its products remain to be investigated in the development and transmission of male gametophyte.Citation59 Arabidopsis genome encodes the homologs of the binding proteins of PtdIns(3,5)P2 and PtdIns3P, such as Arabidopsis β-propellers that binds phosphoinositides, epsin-like protein EpsinR2, and sorting nexin 2b, however, their function is still unknown in plant.Citation60 Further investigations on PtdIns3P metabolism will provide a deeper insight for the function of phosphoinositides in plant reproductive development.
Acknowledgments
This work is funded by the Major Research Plan from the Ministry of Science and Technology of China (No. 2007CB947600) and the National Natural Science Foundation of China (No. 31170293).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Voigt B, Timmers AC, Šamaj J, Hlavacka A, Ueda T, Preuss M, et al. Actin-based motility of endosomes is linked to the polar tip growth of root hairs. Eur J Cell Biol 2005; 84:609 - 21; http://dx.doi.org/10.1016/j.ejcb.2004.12.029; PMID: 16032929
- Vermeer JEM, van Leeuwen W, Tobeña-Santamaria R, Laxalt AM, Jones DR, Divecha N, et al. Visualization of PtdIns3P dynamics in living plant cells. Plant J 2006; 47:687 - 700; http://dx.doi.org/10.1111/j.1365-313X.2006.02830.x; PMID: 16856980
- Jung JY, Kim YW, Kwak JM, Hwang JU, Young J, Schroeder JI, et al. Phosphatidylinositol 3- and 4-phosphate are required for normal stomatal movements. Plant Cell 2002; 14:2399 - 412; http://dx.doi.org/10.1105/tpc.004143; PMID: 12368494
- Park KY, Jung JY, Park J, Hwang JU, Kim YW, Hwang I, et al. A role for phosphatidylinositol 3-phosphate in abscisic acid-induced reactive oxygen species generation in guard cells. Plant Physiol 2003; 132:92 - 8; http://dx.doi.org/10.1104/pp.102.016964; PMID: 12746515
- Leshem Y, Seri L, Levine A. Induction of phosphatidylinositol 3-kinase-mediated endocytosis by salt stress leads to intracellular production of reactive oxygen species and salt tolerance. Plant J 2007; 51:185 - 97; http://dx.doi.org/10.1111/j.1365-313X.2007.03134.x; PMID: 17521408
- Choi Y, Lee Y, Jeon BW, Staiger CJ, Lee Y. Phosphatidylinositol 3- and 4-phosphate modulate actin filament reorganization in guard cells of day flower. Plant Cell Environ 2008; 31:366 - 77; http://dx.doi.org/10.1111/j.1365-3040.2007.01769.x; PMID: 18088331
- Lee Y, Bak G, Choi Y, Chuang WI, Cho HT, Lee Y. Roles of phosphatidylinositol 3-kinase in root hair growth. Plant Physiol 2008; 147:624 - 35; http://dx.doi.org/10.1104/pp.108.117341; PMID: 18408046
- Lee Y, Kim ES, Choi Y, Hwang I, Staiger CJ, Chung YY, et al. The Arabidopsis phosphatidylinositol 3-kinase is important for pollen development. Plant Physiol 2008; 147:1886 - 97; http://dx.doi.org/10.1104/pp.108.121590; PMID: 18515640
- Zhang Y, McCormick S. The regulation of vesicle trafficking by small GTPases and phospholipids during pollen tube growth. Sex Plant Reprod 2010; 23:87 - 93; http://dx.doi.org/10.1007/s00497-009-0118-z; PMID: 20490965
- Chapman LA, Goring DR. Misregulation of phosphoinositides in Arabidopsis thaliana decreases pollen hydration and maternal fertility. Sex Plant Reprod 2011; 24:319 - 26; http://dx.doi.org/10.1007/s00497-011-0172-1; PMID: 21691764
- Xu N, Gao XQ, Zhao XY, Zhu DZ, Zhou LZ, Zhang XS. Arabidopsis AtVPS15 is essential for pollen development and germination through modulating phosphatidylinositol 3-phosphate formation. Plant Mol Biol 2011; 77:251 - 60; http://dx.doi.org/10.1007/s11103-011-9806-9; PMID: 21833541
- Zhang Y, Li S, Zhou LZ, Fox E, Pao J, Sun W, et al. Overexpression of Arabidopsis thaliana PTEN caused accumulation of autophagic bodies in pollen tubes by disrupting phosphatidylinositol 3-phosphate dynamics. Plant J 2011; 68:1081 - 92; http://dx.doi.org/10.1111/j.1365-313X.2011.04761.x; PMID: 21883549
- Herman PK, Emr SD. Characterization of VPS34, a gene required for vacuolar protein sorting and vacuole segregation in Saccharomyces cerevisiae. Mol Cell Biol 1990; 10:6742 - 54; PMID: 2247081
- Mizushima N. Autophagy: process and function. Genes Dev 2007; 21:2861 - 73; http://dx.doi.org/10.1101/gad.1599207; PMID: 18006683
- Funderburk SF, Wang QJ, Yue Z. The Beclin 1-VPS34 complex--at the crossroads of autophagy and beyond. Trends Cell Biol 2010; 20:355 - 62; http://dx.doi.org/10.1016/j.tcb.2010.03.002; PMID: 20356743
- Stack JH, Herman PK, Schu PV, Emr SD. A membrane-associated complex containing the Vps15 protein kinase and the Vps34 PI 3-kinase is essential for protein sorting to the yeast lysosome-like vacuole. EMBO J 1993; 12:2195 - 204; PMID: 8387919
- Budovskaya YV, Hama H, DeWald DB, Herman PK. The C terminus of the Vps34p phosphoinositide 3-kinase is necessary and sufficient for the interaction with the Vps15p protein kinase. J Biol Chem 2002; 277:287 - 94; http://dx.doi.org/10.1074/jbc.M109263200; PMID: 11689570
- Yan Y, Backer JM. Regulation of class III (Vps34) PI3Ks. Biochem Soc Trans 2007; 35:239 - 41; http://dx.doi.org/10.1042/BST0350242; PMID: 17371248
- Itakura E, Kishi C, Inoue K, Mizushima N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell 2008; 19:5360 - 72; http://dx.doi.org/10.1091/mbc.E08-01-0080; PMID: 18843052
- Cao Y, Klionsky DJ. Physiological functions of Atg6/Beclin 1: a unique autophagy-related protein. Cell Res 2007; 17:839 - 49; http://dx.doi.org/10.1038/cr.2007.78; PMID: 17893711
- Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol 2009; 10:458 - 67; http://dx.doi.org/10.1038/nrm2708; PMID: 19491929
- Obara K, Sekito T, Ohsumi Y. Assortment of phosphatidylinositol 3-kinase complexes--Atg14p directs association of complex I to the pre-autophagosomal structure in Saccharomyces cerevisiae. Mol Biol Cell 2006; 17:1527 - 39; http://dx.doi.org/10.1091/mbc.E05-09-0841; PMID: 16421251
- Kihara A, Noda T, Ishihara N, Ohsumi Y. Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J Cell Biol 2001; 152:519 - 30; http://dx.doi.org/10.1083/jcb.152.3.519; PMID: 11157979
- Fujiki Y, Yoshimoto K, Ohsumi Y. An Arabidopsis homolog of yeast ATG6/VPS30 is essential for pollen germination. Plant Physiol 2007; 143:1132 - 9; http://dx.doi.org/10.1104/pp.106.093864; PMID: 17259285
- Qin G, Ma Z, Zhang L, Xing S, Hou X, Deng J, et al. Arabidopsis AtBECLIN 1/AtAtg6/AtVps30 is essential for pollen germination and plant development. Cell Res 2007; 17:249 - 63; PMID: 17339883
- Harrison-Lowe NJ, Olsen LJ. Autophagy Protein 6 (ATG6) is required for pollen germination in Arabidopsis thaliana. Autophagy 2008; 4:339 - 48; PMID: 18227644
- Taylor GS, Maehama T, Dixon JE. Myotubularin, a protein tyrosine phosphatase mutated in myotubular myopathy, dephosphorylates the lipid second messenger, phosphatidylinositol 3-phosphate. Proc Natl Acad Sci U S A 2000; 97:8910 - 5; http://dx.doi.org/10.1073/pnas.160255697; PMID: 10900271
- Walker DM, Urbe´ S, Dove SK, Tenza D, Raposo G, Clague MJ. Characterization of MTMR3. an inositol lipid 3-phosphatase with novel substrate specificity. Curr Biol 2001; 11:1600 - 5; http://dx.doi.org/10.1016/S0960-9822(01)00501-2; PMID: 11676921
- Clague MJ, Lorenzo O. The myotubularin family of lipid phosphatases. Traffic 2005; 6:1063 - 9; http://dx.doi.org/10.1111/j.1600-0854.2005.00338.x; PMID: 16262718
- Parrish WR, Stefan CJ, Emr SD. Essential role for the myotubularin-related phosphatase Ymr1p and the synaptojanin-like phosphatases Sjl2p and Sjl3p in regulation of phosphatidylinositol 3-phosphate in yeast. Mol Biol Cell 2004; 15:3567 - 79; http://dx.doi.org/10.1091/mbc.E04-03-0209; PMID: 15169871
- Robinson FL, Dixon JE. Myotubularin phosphatases: policing 3-phosphoinositides. Trends Cell Biol 2006; 16:403 - 12; http://dx.doi.org/10.1016/j.tcb.2006.06.001; PMID: 16828287
- Ding Y, Lapko H, Ndamukong I, Xia Y, Al-Abdallat A, Lalithambika S, et al. The Arabidopsis chromatin modifier ATX1, the myotubularin-like AtMTM and the response to drought. Plant Signal Behav 2009; 4:1049 - 58; http://dx.doi.org/10.4161/psb.4.11.10103; PMID: 19901554
- Kerk D, Moorhead GBG. A phylogenetic survey of myotubularin genes of eukaryotes: distribution, protein structure, evolution, and gene expression. BMC Evol Biol 2010; 10:196; http://dx.doi.org/10.1186/1471-2148-10-196; PMID: 20576132
- Maehama T, Dixon JE. The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J Biol Chem 1998; 273:13375 - 8; http://dx.doi.org/10.1074/jbc.273.22.13375; PMID: 9593664
- Ueno T, Sato W, Horie Y, Komatsu M, Tanida I, Yoshida M, et al. Loss of Pten, a tumor suppressor, causes the strong inhibition of autophagy without affecting LC3 lipidation. Autophagy 2008; 4:692 - 700; PMID: 18424911
- Gupta R, Ting JT, Sokolov LN, Johnson SA, Luan S. A tumor suppressor homolog, AtPTEN1, is essential for pollen development in Arabidopsis. Plant Cell 2002; 14:2495 - 507; http://dx.doi.org/10.1105/tpc.005702; PMID: 12368500
- Sormani R, Pribat A, Rousseau M, Taconnat L, Renou JP, Meyer C, et al. Over expression of a plant homolog of the human tumor suppressor PTEN leads to flower sterility. 20TH INTERNATIONAL CONFERENCE ON ARABIDOPSIS RESEARCH. 2009; 114.
- Dove SK, Dong K, Kobayashi T, Williams FK, Michell RH. Phosphatidylinositol 3,5-bisphosphate and Fab1p/PIKfyve underPPIn endo-lysosome function. Biochem J 2009; 419:1 - 13; http://dx.doi.org/10.1042/BJ20081950; PMID: 19272020
- Shisheva A. PIKfyve: Partners, significance, debates and paradoxes. Cell Biol Int 2008; 32:591 - 604; http://dx.doi.org/10.1016/j.cellbi.2008.01.006; PMID: 18304842
- Odorizzi G, Babst M, Emr SD. Fab1p PtdIns(3)P 5-kinase function essential for protein sorting in the multivesicular body. Cell 1998; 95:847 - 58; http://dx.doi.org/10.1016/S0092-8674(00)81707-9; PMID: 9865702
- Rutherford AC, Traer C, Wassmer T, Pattni K, Bujny MV, Carlton JG, et al. The mammalian phosphatidylinositol 3-phosphate 5-kinase (PIKfyve) regulates endosome-to-TGN retrograde transport. J Cell Sci 2006; 119:3944 - 57; http://dx.doi.org/10.1242/jcs.03153; PMID: 16954148
- Mueller-Roeber B, Pical C. Inositol phospholipid metabolism in Arabidopsis. Characterized and putative isoforms of inositol phospholipid kinase and phosphoinositide-specific phospholipase C. Plant Physiol 2002; 130:22 - 46; http://dx.doi.org/10.1104/pp.004770; PMID: 12226484
- Whitley P, Hinz S, Doughty J. Arabidopsis FAB1/PIKfyve proteins are essential for development of viable pollen. Plant Physiol 2009; 151:1812 - 22; http://dx.doi.org/10.1104/pp.109.146159; PMID: 19846542
- Hirano T, Matsuzawa T, Takegawa K, Sato MH. Loss-of-function and gain-of-function mutations in FAB1A/B impair endomembrane homeostasis, conferring pleiotropic developmental abnormalities in Arabidopsis. Plant Physiol 2011; 155:797 - 807; http://dx.doi.org/10.1104/pp.110.167981; PMID: 21173023
- Zonia L, Munnik T. Vesicle trafficking dynamics and visualization of zones of exocytosis and endocytosis in tobacco pollen tubes. J Exp Bot 2008; 59:861 - 73; http://dx.doi.org/10.1093/jxb/ern007; PMID: 18304978
- Jin N, Chow CY, Liu L, Zolov SN, Bronson R, Davisson M, et al. VAC14 nucleates a protein complex essential for the acute interconversion of PI3P and PI(3,5)P(2) in yeast and mouse. EMBO J 2008; 27:3221 - 34; http://dx.doi.org/10.1038/emboj.2008.248; PMID: 19037259
- Michell RH, Dove SK. A protein complex that regulates PtdIns(3,5)P2 levels. EMBO J 2009; 28:86 - 7; http://dx.doi.org/10.1038/emboj.2008.270; PMID: 19158662
- Duex JE, Tang F, Weisman LS. The Vac14p-Fig4p complex acts independently of Vac7p and couples PI3,5P2 synthesis and turnover. J Cell Biol 2006; b 172:693 - 704; http://dx.doi.org/10.1083/jcb.200512105; PMID: 16492811
- Zhong R, Burk DH, Nairn CJ, Wood-Jones A, Morrison WH 3rd, Ye ZH. Mutation of SAC1, an Arabidopsis SAC domain phosphoinositide phosphatase, causes alterations in cell morphogenesis, cell wall synthesis, and actin organization. Plant Cell 2005; 17:1449 - 66; http://dx.doi.org/10.1105/tpc.105.031377; PMID: 15805481
- Duex JE, Nau JJ, Kauffman EJ, Weisman LS. Phosphoinositide 5-phosphatase Fig 4p is required for both acute rise and subsequent fall in stress-induced phosphatidylinositol 3,5-bisphosphate levels. Eukaryot Cell 2006; a 5:723 - 31; http://dx.doi.org/10.1128/EC.5.4.723-731.2006; PMID: 16607019
- Hughes WE, Cooke FT, Parker PJ. Sac phosphatase domain proteins. Biochem J 2000; 350:337 - 52; http://dx.doi.org/10.1042/0264-6021:3500337; PMID: 10947947
- Chow CY, Zhang Y, Dowling JJ, Jin N, Adamska M, Shiga K, et al. Mutation of FIG4 causes neurodegeneration in the pale tremor mouse and patients with CMT4J. Nature 2007; 448:68 - 72; http://dx.doi.org/10.1038/nature05876; PMID: 17572665
- Zhang X, Chow CY, Sahenk Z, Shy ME, Meisler MH, Li J. Mutation of FIG4 causes a rapidly progressive, asymmetric neuronal degeneration. Brain 2008; 131:1990 - 2001; http://dx.doi.org/10.1093/brain/awn114; PMID: 18556664
- Zhong R, Ye ZH. The SAC domain-containing protein gene family in Arabidopsis. Plant Physiol 2003; 132:544 - 55; http://dx.doi.org/10.1104/pp.103.021444; PMID: 12805586
- Williams ME, Torabinejad J, Cohick E, Parker K, Drake EJ, Thompson JE, et al. Mutations in the Arabidopsis phosphoinositide phosphatase gene SAC9 lead to overaccumulation of PtdIns(4,5)P2 and constitutive expression of the stress-response pathway. Plant Physiol 2005; 138:686 - 700; http://dx.doi.org/10.1104/pp.105.061317; PMID: 15923324
- Thole JM, Vermeer JE, Zhang Y, Gadella TW Jr., Nielsen E. Root hair defective4 encodes a phosphatidylinositol-4-phosphate phosphatase required for proper root hair development in Arabidopsis thaliana. Plant Cell 2008; 20:381 - 95; http://dx.doi.org/10.1105/tpc.107.054304; PMID: 18281508
- Despres B, Bouissonnie´ F, Wu HJ, Gomord V, Guilleminot J, Grellet F, et al. Three SAC1-like genes show overlapping patterns of expression in Arabidopsis but are remarkably silent during embryo development. Plant J 2003; 34:293 - 306; http://dx.doi.org/10.1046/j.1365-313X.2003.01720.x; PMID: 12713536
- Ton J, Jakab G, Toquin V, Flors V, Iavicoli A, Maeder MN, et al. Dissecting the beta-aminobutyric acid-induced priming phenomenon in Arabidopsis. Plant Cell 2005; 17:987 - 99; http://dx.doi.org/10.1105/tpc.104.029728; PMID: 15722464
- Xue HW, Chen X, Mei Y. Function and regulation of phospholipid signalling in plants. Biochem J 2009; 421:145 - 56; http://dx.doi.org/10.1042/BJ20090300; PMID: 19552624
- Munnik T, Vermeer JE. Osmotic stress-induced phosphoinositide and inositol phosphate signalling in plants. Plant Cell Environ 2010; 33:655 - 69; http://dx.doi.org/10.1111/j.1365-3040.2009.02097.x; PMID: 20429089
- Welters P, Takegawa K, Emr SD, Chrispeels MJ. AtVPS34, a phosphatidylinositol 3-kinase of Arabidopsis thaliana, is an essential protein with homology to a calcium-dependent lipid binding domain. Proc Natl Acad Sci U S A 1994; 91:11398 - 402; http://dx.doi.org/10.1073/pnas.91.24.11398; PMID: 7972072
- Hanada K, Sawada Y, Kuromori T, Klausnitzer R, Saito K, Toyoda T, et al. Functional compensation of primary and secondary metabolites by duplicate genes in Arabidopsis thaliana. Mol Biol Evol 2011; 28:377 - 82; http://dx.doi.org/10.1093/molbev/msq204; PMID: 20736450
- Xiong Y, Contento AL, Bassham DC. AtATG18a is required for the formation of autophagosomes during nutrient stress and senescence in Arabidopsis thaliana. Plant J 2005; 42:535 - 46; http://dx.doi.org/10.1111/j.1365-313X.2005.02397.x; PMID: 15860012
- Ndamukong I, Jones DR, Lapko H, Divecha N, Avramova Z. Phosphatidylinositol 5-phosphate links dehydration stress to the activity of ARABIDOPSIS TRITHORAX-LIKE factor ATX1. PLoS One 2010; 5:e13396; http://dx.doi.org/10.1371/journal.pone.0013396; PMID: 20967218