Abstract
Abscisic acid (ABA) is an essential hormone that controls plant growth, development and responses to abiotic stresses. ABA signaling is mediated by type 2C protein phosphatases (PP2Cs), including HAB1 and ABI2, which inhibit stress-activated SnRK2 kinases and whose activity is regulated by ABA and ABA receptors. Based on biochemical data and our previously determined crystal structures of ABI2 and the SnRK2.6–HAB1 complex, we present the catalytic mechanism of PP2C and provide new insight into PP2C–SnRK2 interactions and possible roles of other SnRK2 kinases in ABA signaling.
Abscisic acid (ABA) is a vital plant hormone that regulates many important physiological processes, including seed germination and bud dormancy, and mediates plant protective responses against abiotic stresses such as drought, cold and salinity. The ABA signal is transmitted through a conserved core pathway that includes soluble PYR/PYL/RCAR receptors, type 2C protein phosphatases (PP2Cs), and subclass 2 of Snf1-related kinases (SnRK2s).Citation1,Citation2 Biochemical and structural analyses have illustrated how ABA at high concentrations binds the PYR/PYL/RCAR receptors to induce a conformational change in the “gate” and “latch” loops at the entrance of the ABA binding pocket.Citation3-Citation7 These changes allow the receptors to bind the PP2Cs and inhibit their catalytic activity by the receptor gate loop, which inserts its serine residue into the PP2C catalytic cleft.Citation3,Citation4,Citation7 In addition, an exposed tryptophan residue of the PP2C flap domain inserts into the receptors to make a water mediated contact with ABA in the pocket and serves as a molecular lock that stabilizes both the gate and latch of the receptor in the ABA-bound, PP2C-docked conformation.Citation3,Citation4,Citation7 PP2C inhibition allows autoactivation of SnRK2s by autophosphorylation of a central serine residue in their activation loops whose phosphorylation is required for full stabilization of the active kinase conformation and presumably for moving the activation loop out of the substrate binding cleft.Citation8-Citation13 The activated kinases can then transmit the ABA signal by phosphorylating downstream effectors, which include transcription factors, ion channels, and ROS-generating NADPH oxidase.Citation2
In the absence of ABA, PP2Cs are active to inhibit the SnRK2 kinases by a two-step mechanism.Citation14 At low concentrations, PP2Cs catalytically reduce SnRK2 activity to a basal level by dephosphorylating their activation loop phosphoserine residues. At high levels, PP2Cs completely inhibit SnRK2 kinases by forming stable complexes with SnRK2s, in which the catalytic sites of both enzymes directly interact with each other. The kinase activation loop inserts into the PP2C catalytic cleft with the phosphate-acceptor serine residue at the active site while the PP2C flap tryptophan inserts into the kinase substrate binding cleft, thus mimicking the binding surfaces in the PP2C-receptor complexes. In this conformation, the kinase active cleft is blocked by PP2C and is kept in an inactive, wide open conformation that completely abrogates kinase activity.Citation14 In addition to the catalytic site interactions, SnRK2s have a highly negatively charged C-terminus, the so-called ABA boxCitation8 or Domain II,Citation12 that binds a positively charged surface on the back site of the PP2C to stabilize the catalytic site interactions.Citation11,Citation14 The high resolution structures of representatives of all three core signaling proteins in isolation and as complexes have provided a comprehensive understanding of their intricate interactions. Here we discuss aspects of our structural analysis and present additional biochemical data that provide new insights into substrate specificity and catalytic mechanism of PP2Cs as well as PP2C activity in the kinase-phosphatase complex and PP2C interactions with the acidic C-termini of other members of SnRK2 family.
Results and Discussion
HAB1 substrate specificity
PP2Cs have minimal sequence specificity toward short phosphopeptides.Citation15 In contrast, we and others have shown that HAB1 displays remarkable specificity toward the specific phosphorylated residues in the activation loop of SnRK2 kinases within the context of the full-length kinases.Citation10,Citation11,Citation14 extends this analysis to overall SnRK2.6 phosphorylation sites. To monitor autophosphorylation that is resistant to dephosphorylation, we incubated SnRK2.6 with radio-labeled ATP and phosphatase, either calf intestinal phosphatase (CIP) or HAB1. CIP can non-specifically remove most of the phosphorylation of SnRK2.6. In contrast, HAB1, which is less specific toward small substrates than CIP,Citation15 cannot remove the bulk of SnRK2.6 autophosphorylation sites, consistent with HAB1 being unable to remove the majority of phosphorylation sites outside of the activation loop of SnRK2.6.
Figure 1. HAB1 fails to efficiently dephosphorylate the bulk of SnRK2.6 auto-phosphorylation sites. Ten µM SnRK2.6 were incubated with [32P]-γ-ATP and either 10 µM HAB1 (A) or 0.3 units calf intestine phosphatase (CIP) (B) for the indicated amount of time. The SnRK2.6 phosphorylation can be efficiently removed by CIP but not by HAB1. Reactions were terminated by boiling in SDS sample buffer, separated by SDS PAGE, and subjected to autoradiography. (C). Cartoon presentation of the selective SnRK2.6 activation loop dephosphorylation in the HAB1 catalytic cleft. S, Ser/Thr phosphate acceptor residues; P, phosphate.
![Figure 1. HAB1 fails to efficiently dephosphorylate the bulk of SnRK2.6 auto-phosphorylation sites. Ten µM SnRK2.6 were incubated with [32P]-γ-ATP and either 10 µM HAB1 (A) or 0.3 units calf intestine phosphatase (CIP) (B) for the indicated amount of time. The SnRK2.6 phosphorylation can be efficiently removed by CIP but not by HAB1. Reactions were terminated by boiling in SDS sample buffer, separated by SDS PAGE, and subjected to autoradiography. (C). Cartoon presentation of the selective SnRK2.6 activation loop dephosphorylation in the HAB1 catalytic cleft. S, Ser/Thr phosphate acceptor residues; P, phosphate.](/cms/asset/a3569f52-b59b-44e2-9708-f93d9eb36f91/kpsb_a_10919694_f0001.gif)
The inability of HAB1 to remove phosphorylation outside of the SnRK2.6 action loop is consistent with our structural observations. We have determined the PP2C structures in three different states: as isolated proteins,Citation14 in complex with ABA receptors,Citation3,Citation16 and in complex with SnRK2.6 kinase.Citation14 Comparison of these PP2C structures revealed that there is little conformational change (including the catalytic active site) among the different states,Citation14 suggesting that the PP2C structure is relatively rigid. The rigid nature of PP2C would prevent the access of large protein substrates to the active site of PP2C while allowing the access of small and flexible peptide substrates. SnRK2.6 overcomes this rigidity problem of the PP2C by forming a complementary interface with HAB1, where the activation loop of SnRK2.6 inserts deeply into the HAB1 active cleft. In this configuration, the side chain of S175 of SnRK2.6 is directly positioned against the catalytic site ().Citation14 The SnRK2.6–HAB1 complex structure thus explains why HAB1 efficiently removes the phosphate group from S175 but not from other phosphorylated residues in SnRK2.6 ().
HAB1 catalytic mechanism
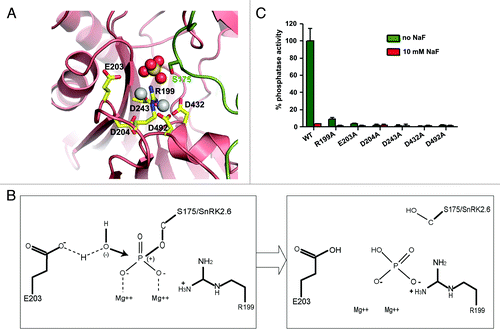
The SnRK2.6–HAB1 interface of the crystal structureCitation14 contains a dominant density for a sulfate group, coordinated by the two Mg2+ ions, resembling the leaving phosphate product of PP2C (). The SnRK2.6–HAB1 structure mimics the post-reaction state of PP2C, and provides a rare opportunity to revisit the catalytic PP2C mechanism (as proposed for mammalian PP2Cα17 and commented on by Dupeux et al.Citation18) in the presence of enzymatic products (). The two Mg2+ ions, which are coordinated by conserved aspartic acids, bind to the phosphate group oxygens of pS175 and thus further increase the partial positive charge of the phosphorus atom. This facilitates the nucleophilic attack of the phosphate by a nearby water molecule, which is deprotonized by the conserved glutamate E203. The reaction is further facilitated by the conserved arginine R199, which binds to the leaving phosphate group by charge interaction and promotes the release of this group from the substrate molecule (SnRK2.6). The importance of each of these residues is supported by mutagenesis (), especially for the four Mg2+ ion-chelating aspartic residues. Conversion of these aspartates to alanines completely inactivated the enzyme (), further supporting the proposed catalytic mechanism above.
Figure 2. PP2C catalytic mechanism. (A) SnRK2.6–HAB1 crystal structure interaction map, in which the sulfate group mimics the phosphate group cleaved from SnRK2.6 S175. This represents the functional correlate of the post-reaction status, at which the phosphate group has been transferred to HAB1 R199. Pink: HAB1, green: SnRK2.6. The sulfate ion is shown in ball presentation, the Mg2+-ions as solid gray balls. (B) Schematic presentation of the reaction mechanism. Mg2+ binds to the phosphate group of the phosphorylated serine, thereby increasing the partial positive charge of the phosphorus atom and facilitating a SN2 nucleophilic attack by the water molecule. E203 partly deprotonizes the water molecule and increases its nucleophilicity for attacking the phosphorus atom. R199 from the PP2C then binds the leaving phosphate group by charge interaction, and removes it from the substrate (SnRK2) molecule. (C) Phosphatase activity of wildtype (WT) HAB1 and HAB1 proteins in which key catalytic residues were altered to alanines. Activities are shown in the absence and presence of 10 mM of the PP2C inhibitor NaF. Error bars indicate SD (n = 3).
SnRK2.6 does not inhibit HAB1
We have demonstrated that the packing of the HAB1 catalytic site against the SnRK2.6 catalytic site completely inhibits SnRK2.6 activity.Citation14 This raises the question whether HAB1 activity is also blocked by SnRK2.6 in the SnRK2.6–HAB1 complex. To test a potential inhibition, we titrated increasing amounts of SnRK2.6 into a phosphatase reaction containing constant amounts of HAB1 and an 11-amino acid phosphopeptide substrate corresponding to the central SnRK2.6 activation loop.Citation19 As shown in , even at 16 µM SnRK2.6 (40-fold molar excess over HAB1), HAB1 activity is unaffected, demonstrating that HAB1 remains catalytically active in the SnRK2.6–HAB1 complex. is a control experiment for , showing both SnRK2.6 and HAB1 are active. Note that this control experiment differs from the dephosphorylation assay () in that SnRK2.6 was preincubated with HAB1 to allow SnRK2.6 inhibition prior to the kinase reaction. In the absence of HAB1, SnRK2.6 is active toward itself and its physiological substrate ABF2. Co-incubation of SnRK2.6 with HAB1, which is present at high concentrations, completely inhibits SnRK2.6 activity. Together, these data demonstrate that HAB1 can completely inhibit SnRK2.6, but SnRK2.6 does not inhibit HAB1, whose inhibition is mediated by ABA receptors.
Figure 3. SnRK2.6 does not inhibit HAB1 activity in the SnRK2.6–HAB1 complex. (A) 0.4 µM HAB1 were preincubated with increasing amounts of SnRK2.6 and then subjected to a phosphatase reaction with 100 µM of a SnRK2.6 activation loop phosphopeptide substrate. Error bars indicate SD (n = 6). (B) Control kinase reaction demonstrating that both SnRK2.6 and HAB1 preparations were active and formed functional complexes. 0.4 µM SnRK2.6 were preincubated with increasing amounts of HAB1 for 30 min and then subjected to a [32P]-γ-ATP kinase reaction.
![Figure 3. SnRK2.6 does not inhibit HAB1 activity in the SnRK2.6–HAB1 complex. (A) 0.4 µM HAB1 were preincubated with increasing amounts of SnRK2.6 and then subjected to a phosphatase reaction with 100 µM of a SnRK2.6 activation loop phosphopeptide substrate. Error bars indicate SD (n = 6). (B) Control kinase reaction demonstrating that both SnRK2.6 and HAB1 preparations were active and formed functional complexes. 0.4 µM SnRK2.6 were preincubated with increasing amounts of HAB1 for 30 min and then subjected to a [32P]-γ-ATP kinase reaction.](/cms/asset/e65bc7f7-6f66-4906-9dfb-b60eea2fbdbe/kpsb_a_10919694_f0003.gif)
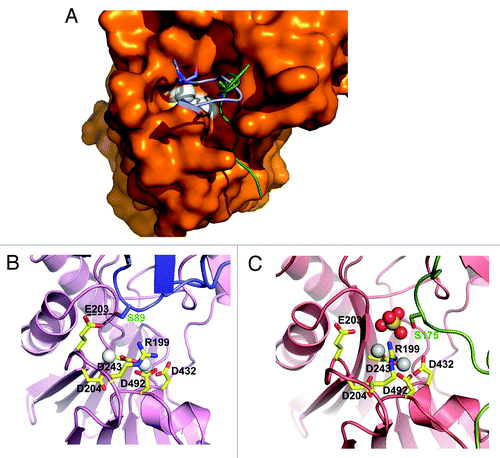
illustrates why binding of the PYL2 gate loop, but not the SnRK2.6 activation loop, in the HAB1 catalytic cleft inhibits HAB1’s phosphatase activity. The gate loop both physically blocks access to the catalytic site () and its S89 hydroxyl group hydrogen-bonds with the catalytic carboxyl group of HAB1 E203 (), so that E203 can no longer deprotonize the catalytic water molecule (see ). The SnRK2.6 activation loop, whose position in the cleft is shifted relative to that of the gate loop (), does not block peptide substrate access to the active site, and the position of its S175 residue is too far from E203 to interfere with the catalytic mechanism. This dual mechanism of PP2C inhibition by ABA-bound PYL2 also explains a seeming discrepancy on the mechanism of HAB1 inhibition. Melcher et al.Citation3 have used a peptide substrate to show that PYL2/ABA can competitively inhibit HAB1. While the isolated peptide in an unfolded linear conformation could enter the catalytic cleft through the channel occupied by the SnRK2.6 activation loop in (see also ref. Citation18), the positions of the activation loop and gate loop peptides clash, consistent with their binding being mutually exclusive. In contrast, Grill and coworkersCitation20,Citation21 demonstrated that PP2Cs are non-competitively inhibited when using the small molecule methyl-umbelliferyl-phosphate (MUP) as substrate. We propose that MUP is small enough to allow binding to the PP2C active site even in the presence of the gate loop, but cannot be dephosphorylated because the gate loop S89 binds the catalytic E203.
Figure 4. Structural mechanism for HAB1 inhibition. (A) Surface presentation of the HAB1 catalytic cleft overlaid with a cartoon presentation of the PYL2 gate loop (blue) and the SnRK2.6 activation loop (green). The gate loop S89 and activation loop S175 are shown as stick presentation. The catalytic Mg2+ ions are indicated as white spheres. (B and C) PYL2 gate loop with S89 in stick presentation (B) and SnRK2.6 activation loop with S175 in stick presentation (C) in the HAB1 catalytic center. Panel C is the same as panel A in and shown for direct comparison.
SnRK2–PP2C interaction is not limited to ABA signaling
Ten members of the SnRK2 family are found in Arabidopsis. The catalytic residues of SnRK2.6 that are involved in HAB1 binding are conserved across this entire family. Although a functional ABA box is only identified in SnRK2.2, 2.3 and 2.6, a high density of negative charge is found at the kinase C-termini of all SnRK2s (). We thus cloned and purified the ABA boxes of all ten SnRK2s to test their interaction with the three PP2Cs ABI1, ABI2, and HAB1. With the exception of SnRK2.1 and 2.8, we clearly detected interaction of all SnRK2 ABA boxes with at least one member of the PP2Cs (). For SnRK2.6–HAB1 it has been shown that both the ABA box and the kinase domain contribute to the full interaction.Citation14 The SnRK2.6 kinase domain residues that directly interact with HAB1 are highly conserved in all SnRK2s (), suggesting that the interactions between PP2Cs and the ABA boxes also occur in the context of full length SnRK2s. While only SnRK2.2, 2.3, and 2.6 are strongly activated by ABA and are clearly positive regulators of ABA signaling,Citation22-Citation24 SnRK2.7 and 2.8 are weakly activated by ABA.Citation25,Citation26 Moreover, a quintuple mutation of the remaining five SnRK2s (2.1/2.4/2.5/2.9/2.10) increased ABA sensitivity in ABA-induced proline accumulationCitation27 and overexpression of a rice homolog of these SnRK2s reduced ABA sensitivity,Citation28 suggesting that some SnRK2s may be negative regulators of ABA signaling.Citation27 In addition to SnRK2s, Ohta et al. demonstrated stable interactions between the PP2C ABI2 and several members of another family of AMPK-related kinases, the PKS/SnRK3s.Citation29 Thus, the observed interactions between multiple SnRKs and PP2Cs suggest their wide-spread co-evolution and co-regulation by mechanisms similar to what has been first observed for SnRK2.6–HAB1. Furthermore, the previously not observed direct interactions of PP2Cs with the C-termini of SnRK2.4, 2.5, 2.7, 2.9, and 2.10 suggest that these kinases may also be involved in ABA signaling.
Figure 5. Both ABA-signaling and non-ABA-signaling SnRK2s interact with PP2Cs. (A) Sequence alignment of the A. thaliana SnRK2 proteins. The residues corresponding to the HAB1-interaction residues of SnRK2.6, including the whole ABA box, are boxed. Negatively charged residues are shown in red, positively charged residues in blue. (B) AlphaScreen interaction between the ABA box regions of the ten A. thaliana SnRK2 proteins fused to H6GST and biotin-MBP-tagged HAB1. Error bars indicate SD (n = 3).
Comparison of the SnRK2.6–HAB1 complex to CDK2–KAP
We recently became aware of another example of a kinase–phosphatase complex, CDK2–kinase-associated phosphatase (KAP),Citation30 in which the catalytic sites of both enzymes are packed against each other in an arrangement very similar to that of SnRK2.6–HAB1 (). In both complexes, the kinase activation loop inserts deeply into the respective phosphatase catalytic cleft. The activation loop residues whose phosphorylation is required for full kinase activity, Ser175 in SnRK2.6 and Thr160 in CDK2, both pack directly against the active phosphatase sites to allow their highly selective dephosphorylation (see above and Poon and HunterCitation31). In addition to their activation loops, the SnRK2.6 and CDK2 kinase domains both also use their αG helices as phosphatase-binding sites. In addition, HAB1 inserts its flap domain (ABA receptor-interaction region) into the catalytic cleft of SnRK2.6 and KAP Lys54 binds CDK2 Tyr15. While the structures of the SnRK2.6 and CDK2 kinase domains are highly similar,Citation13 HAB1 and KAP belong to different classes of phosphatases (PP2Cs and Dual Specificity Phosphatases, respectively) and adopt different positions in their respective complexes relative to the kinase domains ( and S1A).
Figure 6. The SnRK2.6–HAB1 and CDK2–KAP kinase–phosphatase complexes share similar binding characteristics. (A) Cartoon structure of the CDK2–KAP complex (PDB code: 1fq1). KAP is colored brown and CDK2 blue. Mg2+ and the ATP analog AMP-PNP are shown in dot and stick presentation, respectively, in the catalytic cleft of CDK2. P-Thr160 in the CDK2 activation loop is shown in stick presentation. (B) Cartoon structure of the SnRK2.6–HAB1 complex (PDB code: 3UJG). HAB1 is colored red, the HAB1 flap region magenta, and SnRK2.6 green. The catalytic Mg2+ ions in the HAB1 cleft are shown as dots and Ser175 in the SnRK2.6 activation loop in stick presentation. (C) Structural overlay of the CDK2–KAP and SnRK2.6–HAB1 complexes. The same color code is used as in (A) and (B).
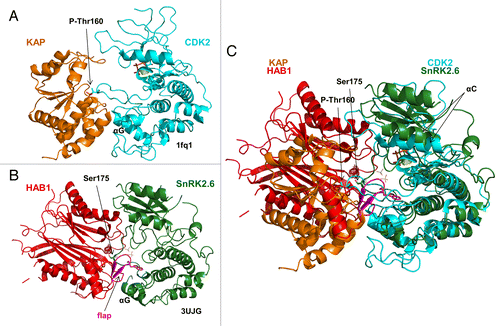
The two complex structures differ in two important points. First, Thr160 is not dephosphorylated in the CDK2–KAP complex, suggesting that KAP may have not been fully active. Second, SnRK2.6 adopted an inactive, wide-open conformation with a blocked catalytic site in its complex with HAB1, consistent with HAB1’s ability to non-catalytically inhibit SnRK2.6.Citation14 In contrast, interaction with KAP induced an active conformation in CDK2Citation30 with the αC helix in the closed conformation and with Mg2+ and the non-cleavable ATP analog AMP-PNP bound in the catalytic cleft (Fig. S1D).
Together, these structures further suggest a possible wide-spread use of kinase–phosphatase complexes with their catalytic sites packed against each other to selectively dephosphorylate activation loop residues. In addition, these interactions may stabilize both active and inactive kinase conformations.
Material and Methods
Protein expression and purification
SnRK2.6 and wildtype and mutant HAB1 were expressed as recombinant H6Sumo and H6GST fusion proteins in E coli and were purified by successive Ni-chromatography, proteolytic tag release, Ni-chromatography, and size-exclusion chromatography as described.Citation14 Recombinant biotinylated HAB1, ABI1, and ABI2 were purified following our published protocol.Citation3 H6GST-ABA box proteins were purified by standard glutathione sepharose chromatography.
SnRK2.6 de-phosphorylation assay
Ten µM SnRK2.6 were incubated with [32P]-γATP and either 10 µM HAB1 or with 0.3 U CIP for the indicated amount of time in 25 mM Tris/Cl, pH 7.3, 5 mM MgCl2, 100 µM EGTA (HAB1 reaction) or 50 mM Tris/Cl, pH 7.5, 1 mM MgCl2 (CIP) at room temperature. Reactions were terminated by boiling in SDS sample buffer and separated by 10% Tricine SDS PAGE. Gels were dried and subjected to autoradiography using a FLA-5000 phosphor imager (Fuji).
Alphascreen interaction assays
Interactions between PP2Cs and ABA box proteins were determined by Alphascreen luminescence proximity assays as described.Citation14 Reactions contained 100 nM recombinant H6GST-ABA box proteins bound to Ni-acceptor beads and 100 nM recombinant biotinylated PP2Cs bound to streptavidin donor beads.
Phosphatase assays
Phosphatase assays were performed by colorimetric determination of phosphate release from phospho-S175 of a SnRK2.6 activation loop phosphopeptide as described previously.Citation3 Reactions contained 200 nM () or 400 nM () recombinant HAB1 protein and 100 µM SnRK2.6 phosphopeptide (HSQPK(pS)TVGTP).
Kinase assays
0.4 µM SnRK2.6 were pre-incubated with the indicated amounts of HAB1 in 25 mM Tris/Cl, pH 7.4, 12 mM MgCl2, and 2 mM DTT for 30 min at room temperature, followed by a 25 min incubation with 0.2 mM unlabeled ATP, 2.5 µCi [32P]-γATP, and 0.2 µM GST-ABF2(73–120) in a total volume of 15 µl. Reactions were terminated by addition of SDS sample buffer and subjected to Tricine SDS-PAGE. Gels were stained with Coomassie and subjected to autoradiography using a FLA-5000 phosphor imager (Fuji).
In vitro mutagenesis
Site-directed mutagenesis was performed using the QuikChange method (Agilent). Mutations and all plasmid constructs were confirmed by sequencing.
Additional material
Download Zip (6.6 MB)Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was supported by the Jay and Betty Van Andel Foundation (H.E.X.) and the Singapore Biomedical Research Council (E.-L.Y.). L.-M.N. and F.-F.S were supported by an overseas PhD scholarship from the NUS Graduate School for Integrative Sciences and Engineering (NGS).
Author Contributions
K.M., and H.E.X. conceived the project and designed research; X.E.Z., F.-F.S., L.-M.N., A.K., K.M.S.-P., K.M. and H.E.X. performed research; F.-F.S., L.-M.N., X.E.Z., A. K., K.M.S.-P, KM, and H.E.X. analyzed data; and K.M. and H.E.X. wrote the paper with contributions from all authors.
References
- Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR. Abscisic acid: emergence of a core signaling network. Annu Rev Plant Biol 2010; 61:651 - 79; http://dx.doi.org/10.1146/annurev-arplant-042809-112122; PMID: 20192755
- Hubbard KE, Nishimura N, Hitomi K, Getzoff ED, Schroeder JI. Early abscisic acid signal transduction mechanisms: newly discovered components and newly emerging questions. Genes Dev 2010; 24:1695 - 708; http://dx.doi.org/10.1101/gad.1953910; PMID: 20713515
- Melcher K, Ng LM, Zhou XE, Soon FF, Xu Y, Suino-Powell KM, et al. A gate-latch-lock mechanism for hormone signalling by abscisic acid receptors. Nature 2009; 462:602 - 8; http://dx.doi.org/10.1038/nature08613; PMID: 19898420
- Miyazono K, Miyakawa T, Sawano Y, Kubota K, Kang HJ, Asano A, et al. Structural basis of abscisic acid signalling. Nature 2009; 462:609 - 14; http://dx.doi.org/10.1038/nature08583; PMID: 19855379
- Nishimura N, Hitomi K, Arvai AS, Rambo RP, Hitomi C, Cutler SR, et al. Structural mechanism of abscisic acid binding and signaling by dimeric PYR1. Science 2009; 326:1373 - 9; http://dx.doi.org/10.1126/science.1181829; PMID: 19933100
- Santiago J, Dupeux F, Round A, Antoni R, Park SY, Jamin M, et al. The abscisic acid receptor PYR1 in complex with abscisic acid. Nature 2009; 462:665 - 8; http://dx.doi.org/10.1038/nature08591; PMID: 19898494
- Yin P, Fan H, Hao Q, Yuan X, Wu D, Pang Y, et al. Structural insights into the mechanism of abscisic acid signaling by PYL proteins. Nat Struct Mol Biol 2009; 16:1230 - 6; http://dx.doi.org/10.1038/nsmb.1730; PMID: 19893533
- Belin C, de Franco PO, Bourbousse C, Chaignepain S, Schmitter JM, Vavasseur A, et al. Identification of features regulating OST1 kinase activity and OST1 function in guard cells. Plant Physiol 2006; 141:1316 - 27; http://dx.doi.org/10.1104/pp.106.079327; PMID: 16766677
- Fujii H, Verslues PE, Zhu JK. Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. Plant Cell 2007; 19:485 - 94; http://dx.doi.org/10.1105/tpc.106.048538; PMID: 17307925
- Umezawa T, Sugiyama N, Mizoguchi M, Hayashi S, Myouga F, Yamaguchi-Shinozaki K, et al. Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc Natl Acad Sci U S A 2009; 106:17588 - 93; http://dx.doi.org/10.1073/pnas.0907095106; PMID: 19805022
- Xie T, Ren R, Zhang YY, Pang Y, Yan C, Gong X, et al. Molecular mechanism for inhibition of a critical component in the Arabidopsis thaliana abscisic acid signal transduction pathways, SnRK2.6, by protein phosphatase ABI1. J Biol Chem 2012; 287:794 - 802; http://dx.doi.org/10.1074/jbc.M111.313106; PMID: 22090030
- Yoshida R, Umezawa T, Mizoguchi T, Takahashi S, Takahashi F, Shinozaki K. The regulatory domain of SRK2E/OST1/SnRK2.6 interacts with ABI1 and integrates abscisic acid (ABA) and osmotic stress signals controlling stomatal closure in Arabidopsis. J Biol Chem 2006; 281:5310 - 8; http://dx.doi.org/10.1074/jbc.M509820200; PMID: 16365038
- Ng LM, Soon FF, Zhou XE, West GM, Kovach A, Suino-Powell KM, et al. Structural basis for basal activity and autoactivation of abscisic acid (ABA) signaling SnRK2 kinases. Proc Natl Acad Sci U S A 2011; 108:21259 - 64; http://dx.doi.org/10.1073/pnas.1118651109; PMID: 22160701
- Soon FF, Ng LM, Zhou XE, West GM, Kovach A, Tan MH, et al. Molecular mimicry regulates ABA signaling by SnRK2 kinases and PP2C phosphatases. Science 2012; 335:85 - 8; http://dx.doi.org/10.1126/science.1215106; PMID: 22116026
- Vlad F, Rubio S, Rodrigues A, Sirichandra C, Belin C, Robert N, et al. Protein phosphatases 2C regulate the activation of the Snf1-related kinase OST1 by abscisic acid in Arabidopsis. Plant Cell 2009; 21:3170 - 84; http://dx.doi.org/10.1105/tpc.109.069179; PMID: 19855047
- Melcher K, Xu Y, Ng LM, Zhou XE, Soon FF, Chinnusamy V, et al. Identification and mechanism of ABA receptor antagonism. Nat Struct Mol Biol 2010; 17:1102 - 8; http://dx.doi.org/10.1038/nsmb.1887; PMID: 20729862
- Das AK, Helps NR, Cohen PT, Barford D. Crystal structure of the protein serine/threonine phosphatase 2C at 2.0 A resolution. EMBO J 1996; 15:6798 - 809; PMID: 9003755
- Dupeux F, Antoni R, Betz K, Santiago J, Gonzalez-Guzman M, Rodriguez L, et al. Modulation of abscisic acid signaling in vivo by an engineered receptor-insensitive protein phosphatase type 2C allele. Plant Physiol 2011; 156:106 - 16; http://dx.doi.org/10.1104/pp.110.170894; PMID: 21357183
- Fujii H, Chinnusamy V, Rodrigues A, Rubio S, Antoni R, Park SY, et al. In vitro reconstitution of an abscisic acid signalling pathway. Nature 2009; 462:660 - 4; http://dx.doi.org/10.1038/nature08599; PMID: 19924127
- Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, et al. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 2009; 324:1064 - 8; PMID: 19407143
- Szostkiewicz I, Richter K, Kepka M, Demmel S, Ma Y, Korte A, et al. Closely related receptor complexes differ in their ABA selectivity and sensitivity. Plant J 2010; 61:25 - 35; http://dx.doi.org/10.1111/j.1365-313X.2009.04025.x; PMID: 19769575
- Fujii H, Zhu JK. Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proc Natl Acad Sci U S A 2009; 106:8380 - 5; http://dx.doi.org/10.1073/pnas.0903144106; PMID: 19420218
- Fujita Y, Nakashima K, Yoshida T, Katagiri T, Kidokoro S, Kanamori N, et al. Three SnRK2 protein kinases are the main positive regulators of abscisic acid signaling in response to water stress in Arabidopsis. Plant Cell Physiol 2009; 50:2123 - 32; http://dx.doi.org/10.1093/pcp/pcp147; PMID: 19880399
- Nakashima K, Fujita Y, Kanamori N, Katagiri T, Umezawa T, Kidokoro S, et al. Three Arabidopsis SnRK2 protein kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, involved in ABA signaling are essential for the control of seed development and dormancy. Plant Cell Physiol 2009; 50:1345 - 63; http://dx.doi.org/10.1093/pcp/pcp083; PMID: 19541597
- Boudsocq M, Barbier-Brygoo H, Laurière C. Identification of nine sucrose nonfermenting 1-related protein kinases 2 activated by hyperosmotic and saline stresses in Arabidopsis thaliana. J Biol Chem 2004; 279:41758 - 66; http://dx.doi.org/10.1074/jbc.M405259200; PMID: 15292193
- Furihata T, Maruyama K, Fujita Y, Umezawa T, Yoshida R, Shinozaki K, et al. Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proc Natl Acad Sci U S A 2006; 103:1988 - 93; http://dx.doi.org/10.1073/pnas.0505667103; PMID: 16446457
- Fujii H, Verslues PE, Zhu JK. Arabidopsis decuple mutant reveals the importance of SnRK2 kinases in osmotic stress responses in vivo. Proc Natl Acad Sci U S A 2011; 108:1717 - 22; http://dx.doi.org/10.1073/pnas.1018367108; PMID: 21220313
- Chae MJ, Lee JS, Nam MH, Cho K, Hong JY, Yi SA, et al. A rice dehydration-inducible SNF1-related protein kinase 2 phosphorylates an abscisic acid responsive element-binding factor and associates with ABA signaling. Plant Mol Biol 2007; 63:151 - 69; http://dx.doi.org/10.1007/s11103-006-9079-x; PMID: 16977424
- Ohta M, Guo Y, Halfter U, Zhu JK. A novel domain in the protein kinase SOS2 mediates interaction with the protein phosphatase 2C ABI2. Proc Natl Acad Sci U S A 2003; 100:11771 - 6; http://dx.doi.org/10.1073/pnas.2034853100; PMID: 14504388
- Song H, Hanlon N, Brown NR, Noble ME, Johnson LN, Barford D. Phosphoprotein-protein interactions revealed by the crystal structure of kinase-associated phosphatase in complex with phosphoCDK2. Mol Cell 2001; 7:615 - 26; http://dx.doi.org/10.1016/S1097-2765(01)00208-8; PMID: 11463386
- Poon RY, Hunter T. Dephosphorylation of Cdk2 Thr160 by the cyclin-dependent kinase-interacting phosphatase KAP in the absence of cyclin. Science 1995; 270:90 - 3; http://dx.doi.org/10.1126/science.270.5233.90; PMID: 7569954