Abstract
Amino acid metabolism lies at the crossroad between nitrogen assimilation, carbon fixation and secondary metabolism. Because of this central position in plant metabolism, amino acid metabolism is tightly regulated by numerous factors to match both demand from the organs and availability of reduced carbon and inorganic nitrogen. While the amino acid biosynthesis enzymes have been shown to be regulated at the transcriptional and protein levels, the genes involved in amino acid sensing, signal transduction and regulation have not yet been identified. The overexpression of Glutamine Dumper1 leads to a large increase in the amino acid content of the plant and, as we show here, to insensitivity to externally applied amino acids. This phenotype is reminiscent of that of the pig1-1 mutant proposed to display a deregulated metabolism. These data suggest that GDU1 is involved in the regulation of amino acid metabolism and transport. As published previously, the analysis of deletion mutants proves that GDU1's VIMAG domain is important for the function of the protein. The present data show furthermore that other regions participate to this function.
Regulation of Amino Acid Metabolism in Plants
Contrary to animals, plants are able to synthesize all amino acids using ammonium and intermediaries from the carbohydrate metabolism. Amino acid synthesis is thus intimately linked to nitrogen absorption efficiency and photosynthetic activity. The integration of these two parameters is achieved at the level of the regulation of the activity of the nitrate reductase which provides the metabolism with ammonium.Citation1
The various pathways leading to the synthesis of amino acids in plants are quite well described.Citation2 Six main tracks can be identified depending on the starting carbohydrate molecule, which comes from the glycolysis, the tricarboxylic acid cycle or the pentose phosphate pathway (). The regulation of the activity of the biosynthesis branches of the branched-chain amino acids (deriving from oxaloacetate) and of the aromatic amino acids (deriving from phosphoenolpyruvate) has been extensively studied. Importantly, some biosynthetic enzymes have been shown to be positively or negatively regulated by end-products (). As a consequence, plant or cell growth is inhibited by external application of an amino acid that drives a negative feedback effect: this amino acid inhibits the activity of one pathway, resulting in the depletion in the other amino acid(s) produced by this branch.Citation3 Transcriptional regulation by different stresses has been shown for some enzymes of the aromatic amino acid pathwayCitation4,Citation5 and the enzymes involved in the production and degradation of proline.Citation6 Furthermore, experimental evidence shows that the activities of the different pathways are adjusted to one another, and the modification of the activity of one pathway leads to the modification of the content in most of the other amino acids.Citation7,Citation8 Guyer et al.Citation9 also showed a clear enhancement of the accumulation of transcripts for enzymes of several pathways in response to an inhibition of histidine synthesis. A cross-pathway regulation of the metabolism seems then to exist in plant, but it is not yet clear whether this regulation takes place at the transcriptional level or/and at the post-translational level. It can be assumed that this cross-pathway regulation depends on amino acid sensing and signal transduction activities. However, the underlying mechanisms and the genes involved remain to be identified.
Resistance of Two Arabidopsis Mutants to High Concentrations of Amino Acids
Until recently, the mutations or manipulations that lead to modifications of amino acid content in plants concerned genes that encode enzymes of amino acid metabolism. In 2004, two papers described the isolation of mutants showing an increase in amino acid content: pig1-1 and gdu1-1D.Citation10,Citation11 Unfortunately the gene responsible for the recessive pig1-1 mutation has not been identified. The Pig1− plants present an increased amino acid metabolism leading to amino acid over-accumulation and insensitivity to elevated amino acid concentrations in the growth medium. The gdu1-1D mutation is caused by the overexpression of a small membrane protein, GDU1. Gdu1D− plants present higher amino acid content and transport within the plant compared to the wild-type and secrete glutamine by the hydathodes.
The GDU1 protein contains two conserved domains: a membrane segment and the plant-specific “VIMAG” domain, present in the C-terminal part of the protein. We showed recently that the VIMAG domain is necessary for GDU1 to cause the Gdu1D− phenotype when overexpressed in plants.Citation12 The striking similarities between the phenotypes of both pig1-1 and gdu1-1D plants prompted us to compare the resistance of these mutants to toxic concentrations of amino acids. The growth of mutants overexpressing various mutated or truncated versions of GDU1 was also tested in that assay. The log1 plants bear a suppressor G to R mutation in the VIMAG domain of GDU1 in the gdu1-6D background, the ΔGDU1 plants over-express GDU1 lacking the VIMAG domain, and the C-GDU1 plants overexpress only the C-terminal domain of GDU1. These three genotypes do not present the Gdu1− phenotype.Citation12
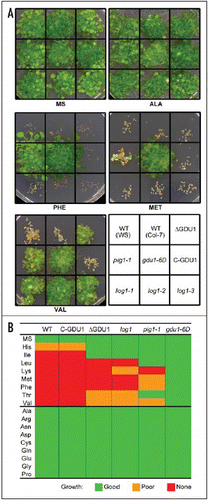
The growth of wild type plants (WS and Col-7 ecotypes) is severely inhibited by the presence of histidine, isoleucine, leucine, lysine, methionine, phenylalanine, threonine and valine (). pig1-1 plants were only susceptible to the branched-chain amino acids leucine, lysine and methionine. This is in good agreement with the fact that this mutant was identified by its resistance to phenylalanine, an aromatic amino acid.Citation10 Strikingly, line gdu1-6D overexpressing the intact GDU1 protein presents no growth inhibition by any of the amino acids in the media, showing that these plants are even less susceptible to amino acids than pig1-1. A reason for this phenotype can be that the plants are less sensitive to the negative feedback effect of amino acids on the metabolic enzymes, or that they present like pig1-1 an increased rate of breakdown and synthesis of amino acids. This is supposed to be a consequence of a deregulation of amino acid homeostasis.Citation10 It is important to note that mutations affecting amino acid transporters can lead to an amino acid resistant phenotype.Citation13–Citation15 Nevertheless, this tolerance is not accompanied by the dramatic changes in amino acid content of the plants seen for pig1-1 and gdu1-6D.Citation10,Citation11
The C-GDU1 plants display no Gdu1− phenotype on soil.Citation12 In good agreement with this fact, these plants behave exactly as the wild type on amino acid containing media. On the contrary, while the plants overexpressing a modified GDU1 protein (log1 and DGDU1) presented no visible phenotype on soil,Citation12 they were found to be resistant to some amino acids (histidine, isoleucine, and valine, and also lysine for the log1 mutants). Despite the loss of most of its functionality by the mutation or the deletion of the VIMAG domain, GDU1 is thus still able to confer resistance to some exogenously applied amino acids. This shows that the VIMAG domain is not the only functional domain of GDU1. The sequence of the transmembrane domain is particularly conserved among GDU proteins,Citation12 suggesting that it could bear part of the function of the protein. The involvement of less conserved, C-terminal stretches of amino acids in the function of GDU1 can however not be excluded.
The first conclusion from the experiment presented here is that although the VIMAG domain is necessary for many aspects of Gdu1− phenotype, other regions of GDU1 are important for the function of the protein. But more importantly, this experiment emphasizes another component of the Gdu1D− phenotype, namely the resistance to externally applied amino acids, which reveals a probable disturbance in the regulation of amino acid metabolism. We propose that the membrane protein GDU1 is involved in the regulation of the amino acid metabolism. GDU1 would be the first gene identified as involved in this process and the study of GDU1 function will help to shed light on an uncharacterized, but central, process of plant physiology.
Figures and Tables
Figure 1 Biochemical pathways for synthesis of amino acids in plants. Starting carbohydrate metabolites are boxed. Thick lines correspond to biochemical pathways and may correspond to more than one reaction. Dotted lines correspond to feedback inhibition (T bar) or activation (arrow) by a metabolite on an enzyme (black dots). (1) chorismate mutase; (2) anthranilate synthase; (3) aspartate kinase; (4) homoserine dehydrogenase; (5) dihydrodipicolinate synthase; (6) threonine synthase; (7) cystathionine gamma-synthase; (8) threonine deaminase; (9) acetohydroxy acid synthase; (10) 2-isopropylmalate synthase. αKG, α-ketoglutarate; PEP, phosphoenolpyruvate; PYR, pyruvate; OAA, oxaloacetate; 3-PG, 3-phosphoglycerate; PRPP, 5-phospho-α-D-ribosyl-1-pyrophosphate; SAM, S-adenosylmethionine. Standard three-letter code is used for amino acids. Adapted from reference Citation2.
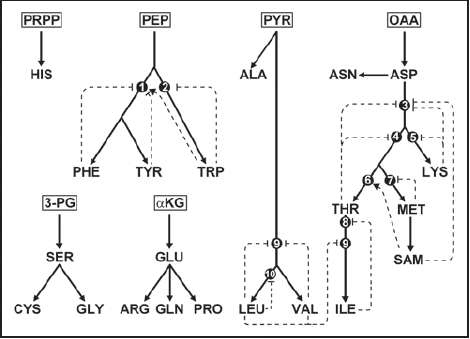
Figure 2 Effect of externally applied amino acids on the growth of pig1-1 and GDU1 overexpressing plants. Seeds were surface sterilized and grown for three weeks on Murashige and Skoog medium (MS, Duchefa) containing 10 mmol/l of the various amino acids (except lysine: 5 mmol/l). (A) Example of growth test for five amino acids. (B) Qualitative summary of the growth on the various media. WS (Wassilewskija) is the wild type corresponding to the pig1-1 mutant while Col-7 is the wild type for all other lines.
Acknowledgements
The authors would like to thank Dr. A. Weber for kindly providing the pig1-1 mutant. The authors are supported by grants from the Deutsche Forschungsgemeinschaft to G.P. (PI 607/1-1 and PI 607/2-1).
References
- Stitt M, Muller C, Matt P, Gibon Y, Carillo P, Morcuende R, Scheible WR, Krapp A. Steps towards an integrated view of nitrogen metabolism. J Exp Bot 2002; 53:959 - 970
- Coruzzi G, Last RL. Buchanan BB, Gruissen W, Jones RL. Amino acids. Biochemistry and Molecular Biology of Plants 2000; Rockville, MD American Society of Plant Physiologists 358 - 410
- Bonner CA, Jensen AB. Recognition of specific patterns of amino acid inhibition of growth in higher plants, uncomplicated by glutamine reversible ‘general amino acid inhibition’. Plant Sci 1997; 130:133 - 143
- Zhao J, Williams CC, Last RL. Induction of Arabidopsis tryptophan pathway enzymes and camalexin by amino acid starvation, oxidative stress, and an abiotic elicitor. Plant Cell 1998; 10:359 - 370
- Zhao J, Last RL. Coordinate regulation of the tryptophan biosynthetic pathway and indolic phytoalexin accumulation in Arabidopsis. Plant Cell 1996; 8:2235 - 2244
- Hare P, Cress W, van Staden J. Review article. Proline synthesis and degradation: A model system for elucidating stress-related signal transduction. J Exp Bot 1999; 50:413 - 434
- Noctor G, Novitskaya L, Lea PJ, Foyer CH. Co-ordination of leaf minor amino acid contents in crop species: Significance and interpretation. J Exp Bot 2002; 53:939 - 945
- Zhu X, Galili G. Increased lysine synthesis coupled with a knockout of its catabolism synergistically boosts lysine content and also transregulates the metabolism of other amino acids in Arabidopsis seeds. Plant Cell 2003; 15:845 - 853
- Guyer D, Patton D, Ward E. Evidence for cross-pathway regulation of metabolic gene expression in plants. Proc Natl Acad Sci USA 1995; 92:4997 - 5000
- Voll LM, Allaire EE, Fiene G, Weber AP. The Arabidopsis phenylalanine insensitive growth mutant exhibits a deregulated amino acid metabolism. Plant Physiol 2004; 136:3058 - 3069
- Pilot G, Stransky H, Bushey DF, Pratelli R, Ludewig U, Wingate VP, Frommer WB. Overexpression of GLUTAMINE DUMPER1 leads to hypersecretion of glutamine from hydathodes of Arabidopsis leaves. Plant Cell 2004; 16:1827 - 1840
- Pratelli R, Pilot G. The plant-specific VIMAG domain of Glutamine Dumper1 is necessary for the function of the protein in arabidopsis. FEBS lett 2006; 580:6961 - 6966
- Lee YH, Fostera J, Chana J, Voll L, Weber A, Tegeder M. AAP1 is functioning in uptake of uncharged amino acids into root cells. Plant J 2007; (in press)
- Hirner A, Ladwig F, Stransky H, Okumoto S, Keinath M, Harms A, Frommer WB, Koch W. Arabidopsis LHT1 is a high-affinity transporter for cellular amino acid uptake in both root epidermis and leaf mesophyll. Plant Cell 2006; 18:1931 - 1946
- Svennerstam H, Ganeteg U, Bellini C, Näsholm T. Comprehensive Screening of Arabidopsis Mutants Suggest the Lysine Histidine Transporter I to be Involved in Plant Uptake of Amino Acids. Plant Physiol 2007; Published on February 9, 2007 http://dx.doi.org/10.1104/pp.106.092205