Abstract
Synthesized by glycogen synthase and starch synthases (SS) using ADP-glucose as the sugar donor molecule, glycogen and starch accumulate as predominant storage carbohydrates in most bacteria and plants, respectively. We have recently shown that the so-called “starch-less” Arabidopsis thaliana adg1–1 and aps1 mutants impaired in ADP-glucose pyrophosphorylase do indeed accumulate low starch content in normal growth conditions, and relatively high starch content when plants were cultured in the presence of microbial volatiles. Our results were strongly supported by data obtained using a highly sensitive method for confocal fluorescence microscopic visualization of iodine stained starch granules. Using Arabidopsis leaves from WT plants, aps1 plants, ss3/ss4 plants lacking both class III and class IV SS, gbss plants lacking the granule-bound SS, and sus1/sus2/sus3/sus4 plants lacking four genes that code for proteins with sucrose synthase activity, in this work we precisely describe the method for preparation of plant samples for starch microscopic examination. Furthermore, we show that this method can be used to visualize glycogen in bacteria, and pure starch granules, amylose and amylopectin.
Starch and glycogen are the most widespread glucose-based reserve polymers in plants and bacteria, respectively. Both are homopolysaccharides of α-1,4-linked glucose subunits with α−1,6-linked glucose at the branched points. Starch accumulates in the form of a quaternary structure composed of two structurally distinct polysaccharides: the highly branched amylopectin (which comprises up to ca. 80% of the starch dry weight) and the infrequently branched amylose. The synthesis of starch requires the participation of starch synthase (SS), which transfers the glucosyl moiety of the activated donor, ADP-glucose, to an elongating glucan chain. Arabidospsis possesses five distinct SS classes: granule-bound SS (GBSS), which is required for the synthesis of amylose, and SS classes I, II, III, and IV, the latter two being suggested to be absolutely required for starch granule initiation.Citation1 Since the initial demonstration that ADP-glucose serves as the precursor molecule for both plant starch and bacterial glycogen biosynthesis,Citation2-Citation4 it became widely considered that ADP-glucose pyrophosphorylase (AGP) is the sole source of ADP-glucose linked to bacterial glycogen and plant starch biosynthesis. In bacteria, genetic evidence that glycogen biosynthesis occurs solely by the AGP pathway has been obtained from the characterization of glgC- mutants impaired in AGP such as the Escherichia coli AC70R1–504 strain.Citation5 These mutants display an apparent glycogen-less phenotype when macroscopically analyzed upon staining with iodine vapors.Citation6 However, recent studies have shown that these mutants can accumulate high levels of glycogen.Citation6 Furthermore, evidence has been provided showing the occurrence of various important sources, other than GlgC, of ADP-glucose linked to glycogen biosynthesis in different bacterial species.Citation7 In Arabidopsis, genetic evidence showing that transitory starch biosynthesis occurs solely by the AGP pathway has been obtained from the characterization of the adg1–1 and aps1 AGP mutants.Citation8-Citation10 Leaves of these mutants display an apparent starch-less phenotype when macroscopically analyzed upon staining with iodine solutions, and when subjected to quantitative-type enzymatic tests for starch measurement. However, during the course of our studies we found that, independently of culture conditions, both adg1–1 and aps1 mutants accumulated ca. 2% of the wild type (WT) starch content despite the total lack of AGP activity and protein in the aps1 mutants.Citation11 Furthermore, leaves of aps1 plants exposed to microbial volatiles emitted by Alternaria alternata accumulated as much as 40% of the starch normally accumulated by illuminated WT leaves.Citation11 Moreover, adg1–1/sex1 and aps1/sex1 double mutants impaired in the machinery required for normal β-amylase-mediated leaf starch mobilization accumulated ca. 3 fold more starch than leaves of adg1–1 and aps1 mutants at the end of the light period.Citation11 Microscopic analyses also provided strong evidence showing the occurrence of transitory starch in leaves of both adg1–1 and aps1 mutants.Citation11 Scanning electron microscopy (SEM) of starch granules isolated from WT and aps1 leaves confirmed not only the presence of small starch granules in aps1 leaves, but also revealed that their shape was comparable to that of starch granules occurring in WT leaves.Citation11 Confocal microscopy analysis of the starch granule marker GBSS–GFP in GBSS–GFP-expressing aps1 and adg1–1 leaves further confirmed the presence of one or two small starch granules in the chloroplasts of adg1–1 and aps1 plants.Citation11
In addition to SEM and GFP confocal fluorescence microscopic localization methods, we developed a sensitive method for confocal fluorescence microscopy detection of starch that allowed to visualize starch granules within the chloroplasts of aps1 and adg1–1 iodine stained leaves.Citation11 In principle, this method is based on the classical iodine staining protocol for macroscopic detection of glycogen and starch.Citation6,Citation12 Leaves from 4-weeks old plants cultured in pots at ambient CO2 (350 ppm) at 20°C under a 16 h light (90 μmol photons sec–1 m−2) / 8 h dark regime were fixed by immersion for 24 h at 37°C into 3.7% formaldehyde and 0.1 M phosphate buffer (pH 6.5). The fixative solution was then washed out with 0.1 M phosphate buffer (pH 6.5) for 24 h at 37°C. Dehydration and decoloration of samples was performed by transferring the samples to 50% (v/v) ethanol for 24 h, and 96% (v/v) ethanol for 2 x 24 h, all steps being conducted at 37°C (at this stage samples can be stored for months at 4°C). Samples were then rehydrated in 50% (v/v) ethanol for 30–60 min, transferred to distilled water for 20–30 min, and stained in iodine solution (2% KI (w/v) and 1% I2 (w/v)) for 60 min in the darkness and at room temperature. Samples were then rinsed gently in distilled water for about 1 min, mounted on microscopic slides, and examined using a D-Eclipse C1 confocal microscope (NIKON, Japan) with Ar 488 nm excitation. Starch granule-specific green fluorescence emission was detected using BA515/30 filter (detector gain setting 7.2), whereas red autofluorescence emission was detected using BA650LP filter (detector gain setting 7.2) (as a reference, GFP fluorescence in GBSS-GFP expressing plantsCitation11 was detected with gain settings ranging between 5 and 6.5). No green fluorescence emission could be detected in samples that were not stained with iodine solution (not shown). Pictures were processed by EZ-C1 software (Gold Ver. 3.40). Cropping and final arrangement of images were made in Adobe Photoshop CS3, Ver. 10.0.1.
Using this method, we could observe large, green fluorescence emitting oval/round structures that were negative for red autofluorescence, and identified as starch granules in bright-field images of WT plants (). Consistent with the presence of reduced starch content in aps1 leaves, the green fluorescence emitted by iodine stained starch granules in chloroplasts of mesophyll cells of aps1 leaves was much smaller than that of WT leaves (). We also analyzed mesophyll cells of the ss3/ss4 mutant impaired in SSIII and SSIV isoforms that are suggested to be absolutely required for starch granule initiation.Citation1 Although Szydlowski et al.Citation1 reported that this double mutant displays a starch-less phenotype, a more recent work has provided evidence that ss3/ss4 leaves accumulate as much as 10% of the WT starch content.Citation13 Consistently, analyses performed using the method for confocal fluorescence microscopic observation of starch granules confirmed the occurrence of starch granules in mesophyll cells of the ss3/ss4 mutant ().
Figure 1. Confocal fluorescence microscopic analysis of iodine stained starch in leaves. Green fluorescence emission of iodine-stained starch granules occurring in chloroplasts of WT, aps1, ss3/ss4, gbss and sus1/sus2/sus3/sus4. Plants were grown in pots at ambient CO2 (350 ppm) at 20°C under a 16 h light (90 μmol photons sec–1 m−2) / 8 h dark regime. Leaves were harvested at the end of the light period, fixed, stained as described in the main text, and examined using a D-Eclipse C1 confocal microscope with Ar 488 nm excitation using BA515/30 filter (detector gain setting 7.2). Note that green fluorescence associated only with oval/round structures that were identified as starch granules. Bar = 5 µm.
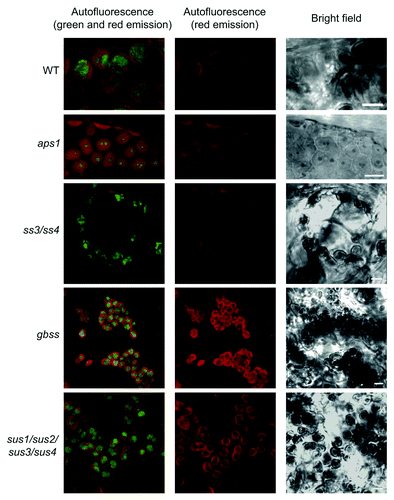
GBSS is involved in the synthesis of amylose, and impairments in this activity result in changes of amylopectin structure.Citation14 Whether the method for confocal fluorescence microscopic observation of starch granules is suitable to visualize starch granules with reduced amylose content was investigated by using a T-DNA gbss Arabidopsis mutant impaired in GBSS activity (GABI_914G01). As shown in these analyses revealed that starch granules from gbss leaves emitted green fluorescence.
In many heterotrophic organs, sucrose synthase (SuSy) activity acts as a major determinant of sink strength that highly controls the conversion of incoming sucrose into starch. SuSy has also been suggested to be involved, at least in part, in the sucrose-starch conversion process in autotrophic organs.Citation15,Citation16 Earlier studiesCitation17,Citation18 have shown that different organs of the sus1/sus2/sus3/sus4 quadruple Arabidopsis mutant accumulate WT starch content. We employed the method for confocal fluorescence microscopic observation described in this work to visualize the starch granules in the chloroplasts of sus1/sus2/sus3/sus4 mutant. As shown in , these analyses revealed the occurrence of large starch granules in the sus1/sus2/sus3/sus4 mesophyll cells, which is consistent with the view that sus1/sus2/sus3/sus4 leaves accumulate nearly WT starch content.
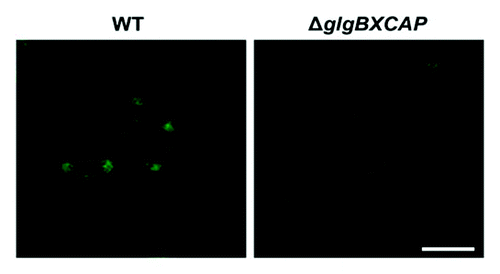
The method for confocal fluorescence microscopic observation of iodine stained branched homopolysaccharides of α-1,4- and α−1,6-linked glucose molecules was valid not only for starch granules occurring in plant cells, but also for glycogen granules occurring in bacteria. E. coli cells cultured in M9 minimal medium (4 mM NaCl, 9 mM NH4Cl, 0.1 mM CaCl2, 2 mM MgSO4, 48 mM Na2HPO4 and 22 mM KH2PO4) supplemented with glucose were harvested at the end of the exponential growth,Citation19 fixed by immersion for 1 h in 3.7% formaldehyde and 0.1 M phosphate buffer (pH 6.5), and centrifuged at 6000 rpm for 5 min. The pellet thus obtained was washed with 0.1 M phosphate buffer and stained in iodine solution (2% KI (w/v) and 1% I2 (w/v)) for 10 min. Stained cells were then transferred to microscopic slides and examined as described above using a D-Eclipse C1 confocal microscope (NIKON, Japan) with Ar 488 nm excitation and green emission detected using BA515/30 filter. As shown in , these analyses revealed the presence in the poles of the cells of fluorescence emitting iodine-stained glycogen, which is consistent with previous electron microscopy studies on topographic distribution of glycogen granules in E. coli.Citation19 In clear contrast, glycogen-less ΔglgBXCAP mutants lacking the whole glycogen biosynthetic machineryCitation20 did not exhibit any green fluorescence.
Figure 2. Confocal fluorescence microscopic observation of iodine stained glycogen granules in E. coli. E. coli cells were cultured in M9 minimal medium supplemented with glucose, harvested at the end of the exponential growth and stained as described in the main text. Note the presence of green fluorescence dots in the poles of WT cells, but not in the glycogen-less ΔglgBXCAP cells lacking the whole glycogen biosynthetic machinery. Bar = 2.5 µm
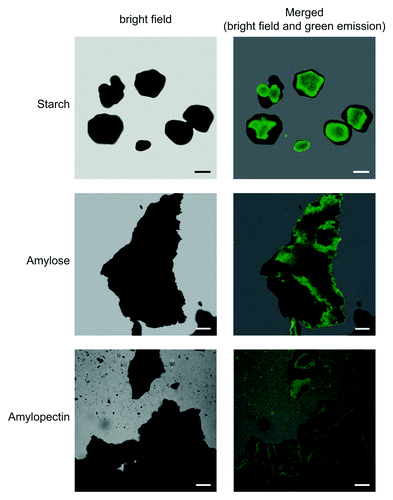
The method for confocal fluorescence microscopic observation of iodine stained starch and glycogen granules was valid not only for structures occurring inside the cell, but also for pure, isolated polymers. Potato starch, amylose and amylopectin were stained with iodine solution (see above), rinsed gently in distilled water for about 1 min, mounted on microscopic slides, and examined using a D-Eclipse C1 confocal microscope with Ar 488 nm excitation. As shown in , in all cases green fluorescence emission was detected using BA515/30 filter (detector gain setting 7.2–7.5). No green fluorescence emission could be detected in samples that were not stained with iodine solution (not shown).
Figure 3. Confocal fluorescence microscopic observation of iodine stained pure potato starch, amylopectin and amylose. Pure, commercially available potato starch (Roche), amylose (Sigma A00512) and amylopectin (Sigma A8515) were stained with iodine solution, rinsed gently in distilled water for about 1 min, mounted on microscopic slides, and examined using a D-Eclipse C1 confocal microscope with Ar 488 nm excitation using BA515/30 filter (detector gain setting 7.2–7.5). Bar = 10 µm for starch granules and 20 µm for amylose and amylopectin.
| Abbreviations: | ||
| AGP | = | ADP-glucose pyrophosphorylase |
| GBSS | = | granule-bound starch synthase |
| GFP | = | green fluorescent protein |
| SEM | = | scanning electron microscopy |
| SS | = | starch synthase |
| SuSy | = | sucrose synthase |
| WT | = | wild type |
Acknowledgments
This research was partially supported by the grants BIO2007–63915 and BIO2010–18239 from the Comisión Interministerial de Ciencia y Tecnología and Fondo Europeo de Desarrollo Regional (Spain), and by Iden Biotechnology S.L.. M.O. was partly supported by grant No. 2/0200/10 from the Grant Academy VEGA. I.E., acknowledges the Spanish Ministry of Culture and Education for a pre-doctoral fellowship. G.A. acknowledges a fellowship from the Public University of Navarre.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Szydlowski N, Ragel P, Raynaud S, Lucas MM, Roldán I, Montero M, et al. Starch granule initiation in Arabidopsis requires the presence of either class IV or class III starch synthases. Plant Cell 2009; 21:2443 - 57; http://dx.doi.org/10.1105/tpc.109.066522; PMID: 19666739
- Recondo E, Dankert M, Leloir LF. Adenosine diphosphate glucose and starch synthesis. Biochem Biophys Res Commun 1963; 12:204 - 7; http://dx.doi.org/10.1016/0006-291X(63)90190-6; PMID: 13973481
- Murata T, Minamikawa T, Akazawa T. Adenosine diphosphate glucose in rice and its role in starch synthesis. Biochem Biophys Res Commun 1963; 13:439 - 43; http://dx.doi.org/10.1016/0006-291X(63)90138-4
- Preiss J, Shen L, Greenberg E, Gentner N. Biosynthesis of bacterial glycogen. IV. Activation and inhibition of the adenosine diphosphate glucose pyrophosphorylase of Escherichia coli B. Biochemistry 1966; 5:1833 - 45; http://dx.doi.org/10.1021/bi00870a008; PMID: 4290042
- Preiss J. Glycogen: biosynthesis and regulation. In, EcoSal—Escherichia coli and Salmonella: Cellular and Molecular Biology. A. Böck, R. Curtiss III, J.B. Kaper, P.D. Karp, F.C. Neidhardt, T. Nyström, J.M. Slauch, C.L. Squires, and D. Ussery, eds (Washington, DC: ASM Press). 2009; Doi:10.1128/ecosal.4.7.4. http://www.ecosal.org.
- Eydallin G, Morán-Zorzano MT, Muñoz FJ, Baroja-Fernández E, Montero M, Alonso-Casajús N, et al. An Escherichia coli mutant producing a truncated inactive form of GlgC synthesizes glycogen: further evidences for the occurrence of various important sources of ADPglucose in enterobacteria. FEBS Lett 2007; 581:4417 - 22; http://dx.doi.org/10.1016/j.febslet.2007.08.016; PMID: 17719034
- Wilson WA, Roach PJ, Montero M, Baroja-Fernández E, Muñoz FJ, Eydallin G, et al. Regulation of glycogen metabolism in yeast and bacteria. FEMS Microbiol Rev 2010; 34:952 - 85; PMID: 20412306
- Wang S-M, Lue W-L, Yu T-S, Long J-H, Wang C-N, Eimert K, et al. Characterization of ADG1, an Arabidopsis locus encoding for ADPG pyrophosphorylase small subunit, demonstrates that the presence of the small subunit is required for large subunit stability. Plant J 1998; 13:63 - 70; http://dx.doi.org/10.1046/j.1365-313X.1998.00009.x; PMID: 9680965
- Sun J, Okita TW, Edwards GE. Modification of carbon partitioning, photosynthetic capacity, and O2 sensitivity in Arabidopsis plants with low ADP-glucose pyrophosphorylase activity. Plant Physiol 1999; 119:267 - 76; http://dx.doi.org/10.1104/pp.119.1.267; PMID: 9880369
- Ventriglia T, Kuhn ML, Ruiz MT, Ribeiro-Pedro M, Valverde F, Ballicora MA, et al. Two Arabidopsis ADP-glucose pyrophosphorylase large subunits (APL1 and APL2) are catalytic. Plant Physiol 2008; 148:65 - 76; http://dx.doi.org/10.1104/pp.108.122846; PMID: 18614708
- Bahaji A, Li J, Ezquer I, Ovecka M, Muñoz FJ, Baroja-Fernández E, et al. Arabidopsis thaliana mutants lacking ADP-glucose pyrophosphorylase can accumulate high levels of starch and ADP-glucose: further evidences for the occurrence of important sources, other than ADP-glucose pyrophosphorylase, of ADP-glucose linked to leaf starch biosynthesis. Plant Cell Physiol 2011; 52:1162 - 76; http://dx.doi.org/10.1093/pcp/pcr067; PMID: 21624897
- Vitha S, Yang M, Sack FD, Kiss JZ. Gravitropism in the starch excess mutant of Arabidopsis thaliana.. Am J Bot 2007; 94:590 - 8; http://dx.doi.org/10.3732/ajb.94.4.590; PMID: 21636428
- Li J, Ezquer I, Bahaji A, Montero M, Ovecka M, Baroja-Fernández E, et al. Microbial volatile-induced accumulation of exceptionally high levels of starch in Arabidopsis leaves is a process involving NTRC and starch synthase classess III and IV. Mol Plant Microbe Interact 2011; 24:1166 - 78; http://dx.doi.org/10.1094/MPMI-05-11-0112
- Delrue B, Fontaine T, Routier F, Decq A, Wieruszeski JM, Van Den Koornhuyse N, et al. Waxy Chlamydomonas reinhardtii: monocellular algal mutants defective in amylose biosynthesis and granule-bound starch synthase activity accumulate a structurally modified amylopectin. J Bacteriol 1992; 174:3612 - 20; PMID: 1592815
- Baroja-Fernández E, Muñoz FJ, Zandueta-Criado A, Morán-Zorzano MT, Viale AM, Alonso-Casajús N, et al. Most of ADP x glucose linked to starch biosynthesis occurs outside the chloroplast in source leaves. Proc Natl Acad Sci U S A 2004; 101:13080 - 5; http://dx.doi.org/10.1073/pnas.0402883101; PMID: 15326306
- Muñoz FJ, Baroja-Fernández E, Morán-Zorzano MT, Viale AM, Etxeberria E, Alonso-Casajús N, et al. Sucrose synthase controls both intracellular ADP glucose levels and transitory starch biosynthesis in source leaves. Plant Cell Physiol 2005; 46:1366 - 76; http://dx.doi.org/10.1093/pcp/pci148; PMID: 15951568
- Barratt DHP, Derbyshire P, Findlay K, Pike M, Wellner N, Lunn J, et al. Normal growth of Arabidopsis requires cytosolic invertase but not sucrose synthase. Proc Natl Acad Sci U S A 2009; 106:13124 - 9; http://dx.doi.org/10.1073/pnas.0900689106; PMID: 19470642
- Baroja-Fernández E, Muñoz FJ, Li J, Bahaji A, Almagro G, Montero M, et al. Sucrose synthase activity in the sus1/sus2/sus3/sus4 Arabidopsis mutant is sufficient to support normal cellulose and starch production. Proc Natl Acad Sci U S A 2012; 109:321 - 6; http://dx.doi.org/10.1073/pnas.1117099109; PMID: 22184213
- Morán-Zorzano MT, Alonso-Casajús N, Muñoz FJ, Viale AM, Baroja-Fernández E, Eydallin G, et al. Occurrence of more than one important source of ADPglucose linked to glycogen biosynthesis in Escherichia coli and Salmonella.. FEBS Lett 2007; 581:4423 - 9; http://dx.doi.org/10.1016/j.febslet.2007.08.017; PMID: 17719035
- Montero M, Almagro G, Eydallin G, Viale AM, Muñoz FJ, Bahaji A, et al. Escherichia coli glycogen genes are organized in a single glgBXCAP transcriptional unit possessing an alternative suboperonic promoter within glgC that directs glgAP expression. Biochem J 2011; 433:107 - 17; http://dx.doi.org/10.1042/BJ20101186; PMID: 21029047