Abstract
Arabidopsis Nup160 and Seh1, encoding two predicted nucleoporins of the Nup107–160 nuclear pore sub-complex, were identified in a reverse genetics screen based on their requirement for basal disease resistance. Both genes also contribute to immunity conferred by Toll interleukin 1 receptor/nucleotide-binding/leucine-rich repeat (TNL)-type R proteins and constitutive resistance activated in the deregulated TNL mutant, snc1. Protein amounts of EDS1, a central regulator of TNL-triggered resistance, are reduced in seh1 and severely depleted in nup160 single mutants. Here, we investigate the impact of mutations in Nup160, Seh1 and a third complex member, MOS3/Nup96, on EDS1 protein accumulation in the snc1 auto-immune mutant background. In addition, we examine the subcellular localization of Seh1 in root tissues.
Plants use sophisticated defense systems against pathogen infection. Cellular resistance signaling involves the transduction of stress stimuli into the nucleus to reprogram the expression of defense genes and subsequent export of transcripts for protein biosynthesis in the cytoplasm to mount an appropriate immune response. The regulated exchange of proteins and RNAs between the cytoplasm and the nucleus is mediated through nuclear pore complexes (NPCs). NPCs are composed of nucleoporins (Nups) that localize to protein sub-complexes within the greater pore.Citation1 The Nup107–160 complex is the largest subunit of the NPC and one of its constituent members in Arabidopsis, MOS3/Nup96, was previously identified in a genetic screen for components that contribute to auto-immunity of the deregulated TNL Resistance (R) gene mutant, snc1 (suppressor of npr1–1, constitutive1).Citation2 Mutations in MOS3 suppress the dwarfism of snc1, its constitutive resistance and increased accumulation of the defense hormone salicylic acid (SA) and pathogenesis-related (PR) gene transcripts. In addition, mos3 single mutants are more susceptible to virulent and avirulent isolates of the bacterial pathogen Pseudomonas syringae and the oomycete Hyaloperonospora arabidopsidis.Citation2 This prompted us to investigate in a reverse-genetics approach whether additional predicted members of the Arabidopsis Nup107–160 complex are required for plant immunity. Our analyses revealed that plants carrying mutations in Nup160 and Seh1 are compromised in basal resistance to virulent Pseudomonas bacteria. Constitutive resistance in snc1 and immunity mediated by other TNL-type R proteins also depend on Nup160 and have a partial requirement for Seh1.Citation3
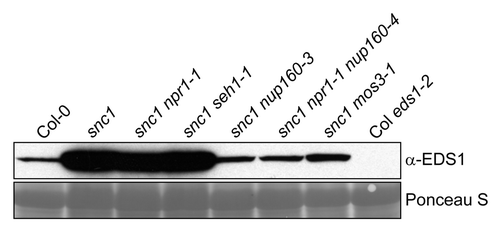
The defense regulator Enhanced Disease Susceptibility1 (EDS1) is indispensable for all known snc1 auto-immune responses,Citation4,Citation5 and although overexpression of EDS1 per se does not cause auto-immunity or enhanced resistance, another molecular phenotype of snc1 plants is the over-accumulation of EDS1.Citation6 Since we found that EDS1 protein amounts are reduced in seh1 and strongly depleted in nup160 single mutant plants,Citation3 we extended our analysis and examined the impact of mutations in Nup160, Seh1 as well as MOS3 on EDS1 protein accumulation in the snc1 auto-immune mutant background. Consistent with the report by García and colleagues,Citation6 our western blot analysis revealed an increased accumulation of EDS1 in snc1 and snc1 npr1–1 compared with Col-0 wild type (). This is probably caused by increased levels of SA and SA-dependent positive feedback expression of EDS1 in snc1.Citation7-Citation9 As compared with snc1, snc1 nup160 and snc1 mos3 double mutant plants harbor wild type-like SA levelsCitation2,Citation3 and, accordingly, total amounts of EDS1 are similar to Col-0 (). In contrast, snc1 seh1 plants still accumulate high levels of SA and resemble the snc1 single mutant in terms of EDS1 protein levels ().Citation3 Nevertheless, seh1 partially suppresses the stunted growth and elevated resistance of snc1, suggesting that Seh1 contributes part of its activity to SA-independent defense responses activated in snc1,Citation5 and reinforcing the notion that increased accumulation of EDS1 is not the sole determinant of constitutive resistance and growth inhibition caused by auto-active snc1.Citation6 Both, nup160 and seh1 are impaired in nuclear mRNA export that may affect EDS1 protein levels.Citation3 While the relatively weak effect of seh1 on EDS1 accumulation may be masked in the EDS1-overaccumulating snc1 background, mutations in Nup160 could have a more pronounced effect on EDS1 abundance in snc1 because Nup160 is not only required for mRNA export but also for full transcriptional expression of the EDS1 gene. This additional function of Nup160 may be required for full SA pathway amplification via the EDS1- and SA-dependent positive feedback loop that is essential for snc1 auto-immunity.Citation7,Citation10
Figure 1. Mutations in Nup160 and MOS3/Nup96 affect EDS1 protein over-accumulation in the snc1 auto-immune mutant. Western blot showing EDS1 levels in total protein extracts of 4-week-old soil-grown plants of the indicated genotypes. Ponceau S staining of the membrane was used to monitor equal loading.
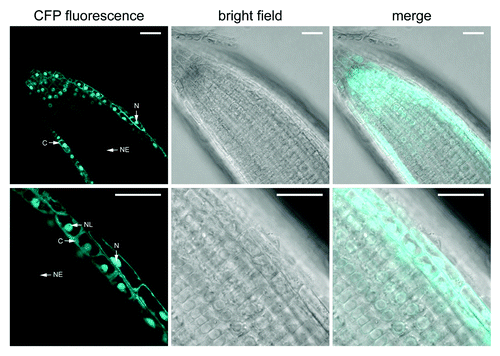
As expected for constituent members of the Nup107–160 complex, MOS3/Nup96 and Nup160 localize to the nuclear rim in root cells of stable transgenic plants that express these proteins as fusions with green fluorescent protein (GFP) under control of the constitutive 35S promoter.Citation2,Citation11 Interestingly, Seh1 fused to cyan fluorescent protein (CFP) shows a nuclear-cytoplasmic distribution when overexpressed in leaf tissues of transgenic plants that complement the enhanced disease susceptibility of seh1–1 () to Pseudomonas syringae pv tomato DC3000.Citation3 This suggests that part of the cellular Seh1 pool is not permanently associated with the Nup107–160 complex. We were unable to detect Seh1-CFP fluorescence by confocal laser scanning microscopy (CLSM) when expressed in transgenic plants by the native Seh1 promoter, likely due to low expression levels. Since the nuclear rim localization of stably overexpressed MOS3-GFP and Nup160-GFP has been examined in roots,Citation2,Citation11 we analyzed the localization of 35S promoter-expressed Seh1-CFP in root tissues of plate-grown transgenic seedlings. Our CLSM analysis of root tips revealed elevated Seh1-CFP fluorescence at the nuclear envelope except in cells of the epidermis and the meristematic zone where Seh1-CFP showed a nuclear and cytoplasmic distribution that was similar to the localization pattern we observed in leaf tissues of these seedlings. This suggests that Seh1 shows tissue-specific differences in its subcellular localization, possibly due to cell-type specific posttranslational modifications that modulate its association with the NPC. However, we cannot exclude the possibility that the overall accumulation of overexpressed Seh1-CFP is higher in leaves as compared with root tissues. This might mask the concentration of CFP fluorescence at the nuclear envelope and thus the stable association of a proportion of Seh1-CFP with the NPC in leaves.
Figure 2. Seh1-CFP subcellular localization in root cells. Confocal images of Seh1-CFP fluorescence in roots of 2-week-old plate-grown seh1–1 seedlings stably expressing Seh1-CFP under control of the double 35S promoter. Scale bars are 25 µm. C, cytoplasm; N, nucleoplasm; NE, nuclear envelope; NL, nucleolus.
Altogether, our data presented here support the notion that Nup160 and Seh1 contribute different activity to TNL-type R protein triggered resistance and auto-immunity in snc1, possibly because of differences in the subcellular localization of both proteins or due to partial compensation of defects in Seh1 by Nup160 or other Nups. Further molecular analyses aim to characterize the detailed mechanisms on how the Nup107–160 complex members Nup160, MOS3 and Seh1 contribute to the regulation of cellular plant immune responses, including those activated in the deregulated TNL R gene mutant, snc1.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We acknowledge funding of our work by the Deutsche Forschungsgemeinschaft (grants WI 3208/4–1 and WI 3208/5–1).
References
- Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, et al. The molecular architecture of the nuclear pore complex. Nature 2007; 450:695 - 701; http://dx.doi.org/10.1038/nature06405; PMID: 18046406
- Zhang Y, Li X. A putative nucleoporin 96 Is required for both basal defense and constitutive resistance responses mediated by suppressor of npr1-1,constitutive 1.. Plant Cell 2005; 17:1306 - 16; http://dx.doi.org/10.1105/tpc.104.029926; PMID: 15772285
- Wiermer M, Cheng YT, Imkampe J, Li M, Wang D, Lipka V, et al. Putative members of the Arabidopsis Nup107-160 nuclear pore sub-complex contribute to pathogen defense. Plant J 2012; 70:796 - 808; http://dx.doi.org/10.1111/j.1365-313X.2012.04928.x; PMID: 22288649
- Li X, Clarke JD, Zhang YL, Dong XN. Activation of an EDS1-mediated R-gene pathway in the snc1 mutant leads to constitutive, NPR1-independent pathogen resistance. Mol Plant Microbe Interact 2001; 14:1131 - 9; http://dx.doi.org/10.1094/MPMI.2001.14.10.1131; PMID: 11605952
- Zhang Y, Goritschnig S, Dong XN, Li X. A gain-of-function mutation in a plant disease resistance gene leads to constitutive activation of downstream signal transduction pathways in suppressor of npr1-1, constitutive 1.. Plant Cell 2003; 15:2636 - 46; http://dx.doi.org/10.1105/tpc.015842; PMID: 14576290
- García AV, Blanvillain-Baufumé S, Huibers RP, Wiermer M, Li G, Gobbato E, et al. Balanced nuclear and cytoplasmic activities of EDS1 are required for a complete plant innate immune response. PLoS Pathog 2010; 6:e1000970; http://dx.doi.org/10.1371/journal.ppat.1000970; PMID: 20617163
- Wiermer M, Feys BJ, Parker JE. Plant immunity: the EDS1 regulatory node. Curr Opin Plant Biol 2005; 8:383 - 9; http://dx.doi.org/10.1016/j.pbi.2005.05.010; PMID: 15939664
- Falk A, Feys BJ, Frost LN, Jones JDG, Daniels MJ, Parker JE. EDS1, an essential component of R gene-mediated disease resistance in Arabidopsis has homology to eukaryotic lipases. Proc Natl Acad Sci U S A 1999; 96:3292 - 7; http://dx.doi.org/10.1073/pnas.96.6.3292; PMID: 10077677
- Jirage D, Tootle TL, Reuber TL, Frost LN, Feys BJ, Parker JE, et al. Arabidopsis thaliana PAD4 encodes a lipase-like gene that is important for salicylic acid signaling. Proc Natl Acad Sci U S A 1999; 96:13583 - 8; http://dx.doi.org/10.1073/pnas.96.23.13583; PMID: 10557364
- Yang SH, Hua J. A haplotype-specific Resistance gene regulated by BONZAI1 mediates temperature-dependent growth control in Arabidopsis. Plant Cell 2004; 16:1060 - 71; http://dx.doi.org/10.1105/tpc.020479; PMID: 15031411
- Dong CH, Hu XY, Tang WP, Zheng XW, Kim YS, Lee BH, et al. A putative Arabidopsis nucleoporin, AtNUP160, is critical for RNA export and required for plant tolerance to cold stress. Mol Cell Biol 2006; 26:9533 - 43; http://dx.doi.org/10.1128/MCB.01063-06; PMID: 17030626