Abstract
PPR (Pentatricopeptide repeat) proteins are mainly involved in RNA metabolism. In Arabidopsis, the PPR family is composed of more than 450 members; however, only few of them were functionally characterized. In a previous report,1 we identified a novel mitochondrial PPR RNA editing factor, named SLO2, which is responsible for 7 editing events in Arabidopsis. Loss-of-function mutation in SLO2 results in plant growth retardation, and delayed development, and leads to the dysfunction of mitochondrial complex I, III and IV. slo2 is the first example of a single gene mutation affecting 3 complexes of the mitochondrial electron transport chain. This Short Communication discusses the conservation of upstream regions of editing sites affected by SLO2 and illustrates the effect of mutation of SLO2 on activation of the alternative pathway. We also reflect upon the implications and perspectives of these findings.
Pentatricopeptide repeat (PPR) protein family which is characterized by tandem arrays of a degenerate 35-amino-acid repeat, is mainly involved in the RNA metabolism, including RNA splicing and editing,Citation2 Based on the protein domains, this family is divided into the P and PLS families. The latter is further divided into PLS, E, E+ and DYW subgroups, based on the C-terminal extension.Citation2 The recently identified PPR protein SLO2 belongs to the E+ subfamily, and acts as the mitochondrial editing factor (MEF).Citation1
Owing to the recently developed SNaPshot technology,Citation3 we were able to identify 7 RNA editing changes in slo2 mutants.Citation1 It was speculated that one MEF may be responsible for editing of multiple sites, as there are more than 400 mitochondrial RNA editing sites as opposed to only about 80 PLS proteins which are predicted to be targeted to mitochondria.Citation2 Thus, our results support this hypothesis. For more than 100 proteins in the E, E+ and DYW subgroups of the PLS subfamily, the subcellular localization could not be unambiguously determined by in silico predictions,Citation2 as was the case for SLO2. Hence, it is very likely that more mitochondrial RNA editing factors will be found in these subgroups.
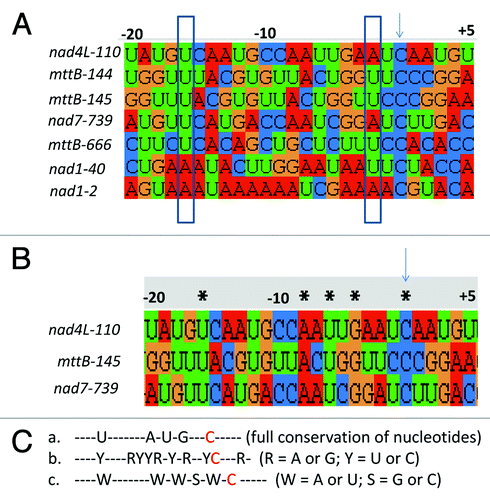
Several studies suggest the presence of cis- elements recognized by RNA editing factors, located approximately 20–25 nucleotides upstream and 1–3 nucleotides downstream of the editing site.Citation4-Citation7 Based on alignment of the target sites of five editing factors involved in editing of multiple sites, it was proposed that editing factors are able to distinguish purines (A and G) from pyrimidines (C and U) and can, at some positions, recognize specific bases. On the other hand, the distinction A/U vs. C/G, based on differences in the number of hydrogen bonding groups in the nucleotide, seems less important in determining binding specificity.Citation8 The putative cis-elements corresponding to 7 editing sites in 4 different RNAs that are affected in slo2 do not share any nucleotide similarity, while two positions show a conservation of the number of intermolecular hydrogen bonding groups ().Citation8 We further compared the putative cis-elements of the three editing sites which are eliminated in slo2 mutants (nad4L-110, mttB-145 and nad7–739), and found that they share only four nucleotides in the upstream region () This degree of identity is much lower than that observed in other examples, such as MEF11 and SLO1 target sites, where 9 and 11 nucleotide identities are shared, respectively.Citation5,Citation6 Although the putative cis-elements of SLO2 targets do not share many nucleotides, several purine-only or pyrimidine-only positions (8 of 20 upstream nucleotides and 1 of 5 downstream nucleotide) are conserved (). Moreover, 5 positions are uniform based on the hydrogen bonding differences, 4 of which with 2 hydrogen bonding groups () .This rises two questions on the possible roles of SLO2: 1, Does SLO2 bind to the cis-element directly? If so, several possibilities exist: first, few key nucleotides may be sufficient in the RNA-recognition process by SLO2; second, purines or pyrimidines may be needed at specific positions; third, the number of hydrogen bonding groups may play a role in the binding specificity. 2, does SLO2 bind to RNA through the intermediate of another RNA editing factor? In that case SLO2 would indirectly bind to the RNA editing sites, and bind to (an) other PPR protein(s) which has(have) the RNA binding activity. Our future research on searching for the SLO2 binding proteins will contribute to solve these questions.
Figure 1. Comparison of putative cis-elements of SLO2 targets. (A) Alignment of putative cis-elements of all RNA editing sites recognized by SLO2. Arrow indicates the editing site, and the rectangles show positions that have a conserved number of hydrogen bonding groups. (B) Alignment of putative cis-elements of the 3 main RNA editing sites recognized by SLO2. * bases conserved among nad4L, mttB and nad7; arrow indicates the editing site. (C) Consensus sequences of the 20 nucleotides upstream of the edited C (red labeled), based on the assumptions proposed previously.Citation8 a, conservation of either one of the 4 nucleotides; b, conservation of purines (R stands for A or G) or pyrimidines (Y stands for U or C). c, conservation based on the number of hydrogen bonding groups (W stands for A or U, S stands for G or C).
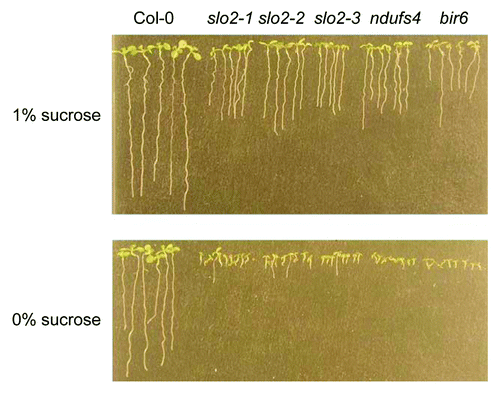
The seeds of slo2 mutants show delayed germination compared with Col-0,Citation1 and slo2 seedling establishment after germination is sugar dependent.Citation1 After the germination stage, Arabidopsis seedlings utilize the storage oils to support their growth. Lack of storage oil in seeds or inefficient conversion from fatty acid to sugar (sucrose, glucose or fructose) through gluconeogenesis could affect seed germination and seedling establishment. Our results indicate that the sugar content is much lower in slo2, and application of various metabolic sugars (glucose, fructose and sucrose) in the medium can help the mutants develop normal roots.Citation1 According to our knowledge, no other RNA editing mutants exhibit such strong phenotype. Interestingly, two complex I related mutants ndufs 4 9 and bir6Citation10 show a similar sugar-dependent phenotype (), suggesting that it may result from the dysfunction of the mitochondrial electron transport chain (mETC), thereby affecting the sugar content. The functional mechanism of the SLO2 protein in this process will be further investigated.
Figure 2.slo2 mutants show a phenotype similar to other complex I mutants on media with and without sucrose. Seeds of Col-0, 3 slo2 alleles, bir6, and ndufs4 were sown on ½ MS medium with or without sucrose. Pictures were taken after 7 d.
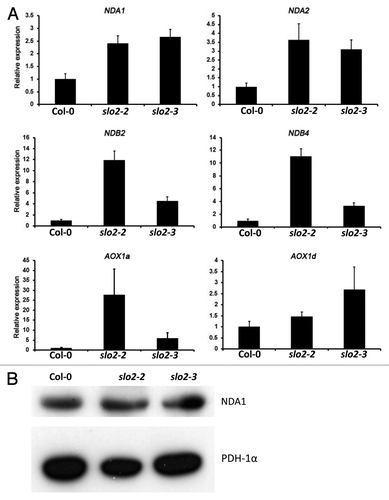
Inhibition of complex I leads to an induction of the alternative pathway.Citation11 To check whether the slo2 mutation leads to changes in the accumulation of transcripts involved in the alternative pathway, we analyzed the gene expression pattern of NDA1, NDA2 (alternative internal NAD(P)H dehydrogenases); NDB2 and NDB4 (alternative external NAD(P)H dehydrogenases); AOX1a and AOX1d (alternative oxidase) by quantitative RT-PCR. All transcripts were significantly induced in slo2 mutants (). These results corroborate our previous observation of enhanced levels of AOX protein in slo2 mutantsCitation1 In order to assess whether the change in transcript level of the alternative NADH dehydrogenase subunit NDA1 affects protein accumulation, we performed a western blot with an anti-NDA1 antibody. While the transcript level was significantly higher in slo2 mutants, the protein abundance was comparable with that of the wild type (). Similar results were observed in the ndufs4 mutant.Citation9 The fact that RNA and protein accumulation do not match was attributed to the fact that NDA1 protein is dually localized in both mitochondria and peroxisomes.Citation9 Hence, loss of function of SLO2 leads to activation of the alternative pathway in Arabidopsis, reflecting an inhibition of the mETC. Although complex I, III and IV are all affected in the slo2 mutants, the plants still can survive; suggesting that compensatory mechanism(s) are induced to produce enough ATP to keep the plant alive. One hypothesis would be an increased flux through the glycolysis and the TCA cycle to increase the amount of ATP produced by substrate level phosphorylation. The NADH produced would be oxidized by the alternative NADH dehydrogenases and the activity of complex II would be increased. This would partially compensate for the absence of complex I by feeding more electrons into the respiratory chain. Therefore, slo2 mutants may act as a good candidate to study the correlations between mitochondrial complex II and other mETC complexes.
Figure 3. Expression of alternative respiratory pathway transcripts in wild type and slo2 mutants. (A) Rosette leaves from 1 mo old plants were harvested and total RNA was extracted. Quantitative real-time PCR were performed using gene specific primers, actin 11 was used as an internal reference. The data shown represent mean values and standard errors obtained from three independent experiments. (B) western blot of NDA1 in mitochondrial fractions from wild type and two slo2 alleles, pyruvate dehydrogenase-1α (PDH-1α) was used as a loading control.
In summary, we identified a novel mitochondrial PPR RNA editing factor, which controls the highest number of RNA editing sites in Arabidopsis ever reported, thereby regulating the carbon energy balance through maintenance of the proper function of the mETC. Our future work will focus on the identification of SLO2 binding partners to unravel its functional mechanism.
| Abbreviations: | ||
| PPR | = | Pentatricopeptide repeat |
| MEF | = | mitochondrial editing factor |
Acknowledgments
Catherine Colas des Francs-Small is gratefully acknowledged for bir6 and ndufs4 seeds. DVDS thanks the Research Foundation Flanders (project G.0313.05) and Ghent University for financial support. EHM thanks the Centre National de la Recherche Scientifique and the Marie Curie Actions (grant PIRG256398) for financial support.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Zhu Q, Dugardeyn J, Zhang C, Takenaka M, Kühn K, Craddock C, et al. SLO2, a mitochondrial pentatricopeptide repeat protein affecting several RNA editing sites, is required for energy metabolism. [Epub ahead of print] Plant J 2012; http://dx.doi.org/10.1111/j.1365-313X.2012.05036.x; PMID: 22540321
- Lurin C, Andrés C, Aubourg S, Bellaoui M, Bitton F, Bruyère C, et al. Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 2004; 16:2089 - 103; http://dx.doi.org/10.1105/tpc.104.022236; PMID: 15269332
- Takenaka M, Brennicke A. Multiplex single-base extension typing to identify nuclear genes required for RNA editing in plant organelles. Nucleic Acids Res 2009; 37:e13; http://dx.doi.org/10.1093/nar/gkn975; PMID: 19059998
- Tang J, Kobayashi K, Suzuki M, Matsumoto S, Muranaka T. The mitochondrial PPR protein LOVASTATIN INSENSITIVE 1 plays regulatory roles in cytosolic and plastidial isoprenoid biosynthesis through RNA editing. Plant J 2010; 61:456 - 66; http://dx.doi.org/10.1111/j.1365-313X.2009.04082.x; PMID: 19929879
- Sung TY, Tseng CC, Hsieh MH. The SLO1 PPR protein is required for RNA editing at multiple sites with similar upstream sequences in Arabidopsis mitochondria. Plant J 2010; 63:499 - 511; http://dx.doi.org/10.1111/j.1365-313X.2010.04258.x; PMID: 20497377
- Verbitskiy D, Zehrmann A, van der Merwe JA, Brennicke A, Takenaka M. The PPR protein encoded by the LOVASTATIN INSENSITIVE 1 gene is involved in RNA editing at three sites in mitochondria of Arabidopsis thaliana. Plant J 2010; 61:446 - 55; http://dx.doi.org/10.1111/j.1365-313X.2009.04076.x; PMID: 19919573
- Takenaka M. MEF9, an E-subclass pentatricopeptide repeat protein, is required for an RNA editing event in the nad7 transcript in mitochondria of Arabidopsis. Plant Physiol 2010; 152:939 - 47
- Hammani K, Okuda K, Tanz SK, Chateigner-Boutin AL, Shikanai T, Small I. A study of new Arabidopsis chloroplast RNA editing mutants reveals general features of editing factors and their target sites. Plant Cell 2009; 21:3686 - 99; http://dx.doi.org/10.1105/tpc.109.071472; PMID: 19934379
- Meyer EH, Tomaz T, Carroll AJ, Estavillo G, Delannoy E, Tanz SK, et al. Remodeled respiration in ndufs4 with low phosphorylation efficiency suppresses Arabidopsis germination and growth and alters control of metabolism at night. Plant Physiol 2009; 151:603 - 19; http://dx.doi.org/10.1104/pp.109.141770; PMID: 19675153
- Koprivova A, des Francs-Small CC, Calder G, Mugford ST, Tanz S, Lee BR, et al. Identification of a pentatricopeptide repeat protein implicated in splicing of intron 1 of mitochondrial nad7 transcripts. J Biol Chem 2010; 285:32192 - 9; http://dx.doi.org/10.1074/jbc.M110.147603; PMID: 20682769
- Moller IM. Plant mitochondria and oxidative stress: electron transport, NADPH turnover, and metabolism of reactive oxygen species. Annu Rev Plant Physiol Plant Mol Biol 2001; 52:561 - 91; http://dx.doi.org/10.1146/annurev.arplant.52.1.561; PMID: 11337409