Abstract
The subcellular polarity of PIN-FORMEDs (PINs) is critical for directional cell-to-cell transport of auxin. Phosphorylation of PIN proteins plays an important role in generating and maintaining specific PIN polarity. In a recent study, we have shown that phosphorylation in certain conserved residues of the PIN3 hydrophilic loop (HL) modulates its subcellular localization and polarity in a cell type-specific manner in different root tissues. Here, we additionally show that the phosphorylation code of PIN3-HL is operational for the determination of PIN3 polarity in the Arabidopsis guard cell and is deciphered in a differential way even in a single tobacco cell for the intracellular trafficking of PIN3. On the other hand, PIN3 localization often remained unaltered in certain cell types irrespective of its phosphorylation status. These findings, together with previous reports, indicate that the phosphorylation code of the PIN-HL along with cell type-specific factors, kinases, and developmental/environmental cues is instrumental for the PIN trafficking to different subcellular compartments as well as different plasma membrane domains.
Protein phosphorylation is generally operational in determination of subcellular polarity both in prokaryotes and eukaryotes.Citation1 In animals, PAR (partition defective) proteins mediate cell polarity and bilateral symmetry, and the phosphorylation of PARs or their targets directly determines polar localization of the target proteins.Citation2,Citation3 Similarly in plants, phosphorylation is implicated in the modulation of polar trafficking of target cargos which is responsible for the directional cell-to-cell transport of the phytohormone auxin and nutrients.Citation1,Citation4 The directional cell-to-cell movement of auxin is driven by a group of transmembrane influx and efflux carrier proteins; AUXIN RESISTANT1 (AUX1)/LIKE-AUX1s for influx and PINs and several ATP-binding cassette-type transporter subfamily B proteins for efflux.Citation1,Citation4 Among these auxin transporters, in particular, the PIN family members play pivotal roles in auxin-related developmental processes with their differential subcellular localization patterns.Citation4
Several recent studies have characterized the phosphorylation sites in the central hydrophilic loop of PIN (PIN-HL) proteins which play a role in determination of PIN polarity.Citation5-Citation10 Our recent study has functionally identified the phosphorylation sites of PIN3-HL that are required for the trafficking and polarity of PIN3 and thus for the biological functions of PIN3.Citation11 The phosphorylation sites (five Ser/Thr residues) in the PIN3-HL are located into two conserved neighboring motifs, RKSNASRRSF(/L) and TPRPSNL (together called as the ‘M3′ motif). The latter is overlapped with one of the formerly known TPRXS motifCitation8 in PIN1 and PIN2 and the former is newly characterized by our study.Citation11 Although these five individual phosphorylation sites are functionally redundant in modulation of PIN3 trafficking, the former RKSNASRRSF(/L) motif (so called ‘3m1’ motif) was found to be more important than its adjacent TPRPSNL motif (2m3) for PIN3 trafficking.Citation11 Wild-type PIN3 localizes in the plasma membrane (PM) of root hair cells in a non-polar manner, whereas it is polarly localized (basal and outer lateral) in the pericycle cell PM.Citation11 In the root hair cell, the phospho-defective PIN3 mutant (M3PIN3, all five Ser/Thr to Ala version) proteins targeted to the tonoplast rather than the PM. Conversely, in the pericycle cell, M3PIN3 targeted normally to the PM but its polarity was lost.Citation11 Intriguingly, in root vascular cells, both polarity and PM localization of M3PIN3 remained unaltered as seen in wild-type PIN3.Citation11
Here, we present additional data showing the role of the M3 motif in PIN3 trafficking. When ectopically expressed in the root epidermal cell using the PIN2 promoter, both wild-type PIN3 and M3PIN3 normally targeted to the PM but with no polarity (). However, when expressed in its own domain under the PIN3 promoter, M3PIN3 lost its polarity in the guard cell. While wild-type PIN3 mainly localized to the outer PM, M3PIN3 mostly resided in both outer and inner PM sides of the guard cell (). These data further support the previous results that the M3 phosphorylation sites of PIN3-HL operate distinctively depending on cell type, indicating that the M3 phosphorylation code is deciphered differentially by distinctive cell type-specific trafficking factors.
Figure 1. Differential intracellular trafficking of phospho-defective PIN3 proteins in diverse cell types. (A) The localization PIN3 and M3PIN3 in the Arabidopsis root epidermal cell. Wild-type PIN3 and M3PIN3 were expressed under the PIN2 promoter. Here, both PIN3 and M3PIN3 showed the similar non-polar PM localization. (B) The localization pattern of PIN3 and M3PIN3 in guard cells of the Arabidopsis cotyledon. Wild-type PIN3 and M3PIN3 were expressed under the PIN3 promoter. M3PIN3 exhibited non-polar localization compared with the polar PIN3 localization in the guard cell PM. (C) The subcellular localization pattern of PIN3, M3PIN3 and 3m1PIN3 in tobacco BY2 cells. Wild-type and mutant PIN3 proteins were induced by 10 µM dexamethason (Dex) for 24 h. PIN3 predominantly localized to the PM, whereas both M3PIN3 and 3m1PIN3 localized to both PM and ER-like compartments pattern as overlapped by the PM/endocytic tracer dye FM4–64 (FM). (D) Localization of PIN3, M3PIN3, and 3m1PIN3 in the cell plate (white arrowheads) of dividing tobacco BY-2 cells. Cells were treated with 25 µM of BFA for 2 h after 24 h Dex induction. All the wild-type and mutant PIN3 proteins were translationally fused with the green fluorescent protein (GFP). Bar = 5 µm for all.
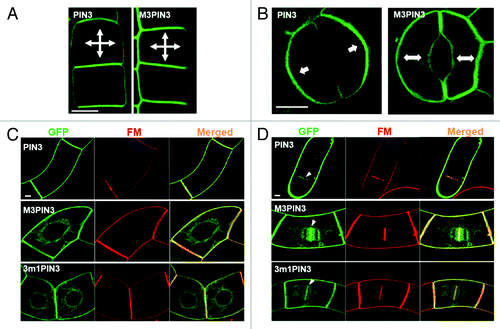
When expressed in tobacco BY-2 cells, PIN3 has been shown to target to the PMCitation12,Citation13 (). Interestingly, both M3PIN3 and 3m1PIN3 were found in the PM as well as in ER-like intracellular compartments of the tobacco cell (). In the brefeldin A (BFA)-treated tobacco cell, wild-type PIN3, M3PIN3, and 3m1PIN3 were all found to localize in the cell plate (), indicating that phospho-defective (M3 and 3m1) PIN3 proteins are also able to localize to the PM and undergo endocytosis similarly with the native PIN3 protein. Furthermore, this localization study with tobacco cells also demonstrates that defects in phosphorylation partially disrupt the capability of PIN3 to target to the PM. This partial role of the PIN3 phosphorylation sites for PM targeting in tobacco cells is contrasted to their major role for PM targeting in Arabidopsis root hair cells.11These data suggest that the phosphorylation code can be read differently in different cell types and even within a single cell type as seen in the tobacco cell case.
Several recent studies on the trafficking of PIN1 and PIN2 have demonstrated that phosphorylation is critical for the polarity determination but not for the PM targeting of PINs.Citation5-Citation10 De novo-synthesized PIN proteins are secreted in a non-polar manner to the PM where they undergo endocytosis and subsequent recycling to generate the polarity.Citation7,Citation12 The poleward recycling of PINs to the apical or the basal targeting pathway depends on their phosphorylation by AGC3 kinases. Closely related AGC3 kinases such as PID, WAG1 and WAG2 directly phosphorylate the serine residues of the TPRXS motifs (three in PIN1–4, and 7 and two in PIN6) in the PIN-HL, resulting in apically targeted PINs (PIN1 and PIN2).Citation7,Citation8 Low or inadequate phosphorylation leads PINs to basal targeting.Citation7,Citation8
In contrast, other recent studies with PIN3 revealed some differences in phosphorylation-mediated trafficking between PIN3 and other PINs. It seems that hyper-phosphorylation of PIN3 by AGC3 kinase members generally causes non-polar PIN3 localization. Increased PID activity in the dark led to apolar PIN3 localization in the hypocotyl endodermis.Citation10 Similarly, INDIHISCENT (the valve margin identity factor in Arabidopsis silique development)-mediated increase in AGC3 kinase (PID and WAG2) expression also resulted in apolar PIN3 localization in root endodermal cells.Citation13 Hence, unlike PIN1 and PIN2 cases where AGC3 kinase-mediated phosphorylation mainly leads to apical localization of those PINs, PIN3 shows predominantly apolar localization under similar conditions. Even though the three TPRXS motifs are highly conserved among PINs, when expressed in the root epidermal cell, PIN1 and PIN2 maintained the apical/basal polarity but PIN3 and PIN4 lost the polarity.Citation14
This distinctive regulation of PIN3 trafficking from those of PIN1 and PIN2 could be due to the distinctive RKSNASRRSF(/L) (3m1) phosphorylation motif in the PIN3-HL.Citation11 The structural context of the 3m1 motif is unique to the PIN347 clade including PIN3, 4, and 7 in that PIN3 does not but PIN1 and PIN2 have a linker between ‘RKSNAS’ and ‘RRSF(L)’Citation11. The structural feature of the 3m1 of the Arabidopsis PIN347 clade is also conserved in other angiosperm species.Citation11 The distinctive phosphorylation code in the 3m1 motif may endow the PIN347 clade proteins with differential trafficking behaviors.
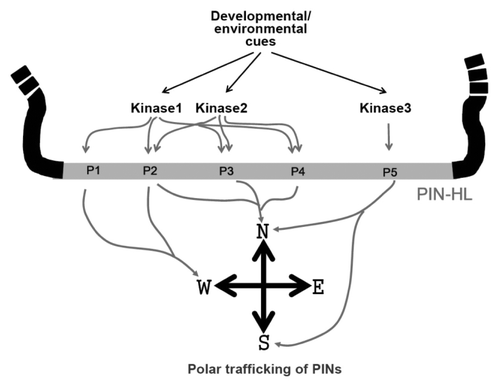
The phosphorylation code for differential trafficking behaviors among PINs should be a synthetic result of distinctive phosphorylation contexts on the PIN-HL, substrate specificity between PINs and kinases, cell type-specific factors translating the phosphorylation code, and developmental/environmental cues regulating spatio-temporal activity of these components (). The mechanistic studies on phosphorylation-mediated trafficking of plant proteins have just begun. The very next challenging task in this subject would be to identify those cell type-specific trafficking factors and to characterize the relationship between the phosphorylation code and the kinases-PINs substrate specificity.
Figure 2. A model illustrating phosphorylation code-mediated modulation of distinctive PIN polarity. The PIN-HL contains multiple conserved phosphorylation motifs (P1~5) on which multiple related kinases (Kinase1~3) act. Phosphorylation of different phosphorylation motifs and/or differential combinatorial phosphorylation among those motifs, which are likely to be targeted by the upstream developmental/environmental cues, may instrument to generate differential poleward trafficking of PINs.
Materials and Methods
The binary vector pCAMBIA1300-NOS with modified cloning sitesCitation15 was used for transgene construction. For the ProPIN3:PIN3-GFP, ProPIN3:M3PIN3-GFP and ProPIN3:3m1PIN3-GFP constructs, construction of the PIN3 promoter, PIN3-GFP, M3PIN3-GFP and 3m1PIN3-GFP, the primer information and cloning sites were described previously.Citation11 For ProTA:PIN3-GFP, ProTA:M3PIN3-GFP and ProTA:3m1PIN8-GFP constructs, the PIN-GFP coding regions were inserted into XhoI-SpeI sites in the modified ProTA7002 vectorCitation16 after restriction enzyme digestion and releasing them from their ProPIN3:PIN-GFP versions with Sal1-Spe1 sites. For the ProPIN2:PIN3-GFP and ProPIN2:M3PIN3-GFP constructs, the 2016-bp PIN2 promoter was amplified with the forward primer 5′-CGA CGT TTA AAC TGC AAG GAT ATC ATT ACC AGT ACC G-3′ and the reverse primer 5′-CTG GTC GAC TTT GAT TTA CTT TTT CCG GCG AGA G-3′ from Arabidopsis genomic DNA and inserted into the pCAMBIA1300-NOS vector (Pme1-Sal1 sites). PIN3-GFP and M3PIN3-GFP inserts were introduced under the PIN2 promoter in the Sal1-Spe1 sites as described above. Tobacco BY-2 cells culture, transformation, Arabidopsis transformation and growth, pharmacological treatments and confocal microscopic observations were described previously.Citation17,Citation18
| Abbreviations: | ||
| BFA | = | brefeldin A |
| ER | = | endoplasmic reticulum |
| GFP | = | green fluorescent protein |
| HL | = | hydrophilic loop |
| PIN | = | PIN-FORMED |
| PM | = | plasma membrane |
| PID | = | PINOID |
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This research was supported by grants from the Mid-career Researcher Program (2011–0017242, NRF, MEST) and the Next-Generation BioGreen 21 programs (TAGC PJ00820701 and SSAC PJ00814102) of the Rural Development Administration. AG was partly supported by a graduate student fellowship from the Korea Research Foundation.
References
- Ganguly A, Sasayama D, Cho HT. Regulation of the polarity of protein trafficking by phosphorylation. Mol Cells 2012; 33:423 - 30; http://dx.doi.org/10.1007/s10059-012-0039-9; PMID: 22453777
- Goldstein B, Macara IG. The PAR proteins: fundamental players in animal cell polarization. Dev Cell 2007; 13:609 - 22; http://dx.doi.org/10.1016/j.devcel.2007.10.007; PMID: 17981131
- Nance J, Zallen JA. Elaborating polarity: PAR proteins and the cytoskeleton. Development 2011; 138:799 - 809; http://dx.doi.org/10.1242/dev.053538; PMID: 21303844
- Grunewald W, Friml J. The march of the PINs: developmental plasticity by dynamic polar targeting in plant cells. EMBO J 2010; 29:2700 - 14; http://dx.doi.org/10.1038/emboj.2010.181; PMID: 20717140
- Michniewicz M, Zago MK, Abas L, Weijers D, Schweighofer A, Meskiene I, et al. Antagonistic regulation of PIN phosphorylation by PP2A and PINOID directs auxin flux. Cell 2007; 130:1044 - 56; http://dx.doi.org/10.1016/j.cell.2007.07.033; PMID: 17889649
- Zourelidou M, Müller I, Willige BC, Nill C, Jikumaru Y, Li H, et al. The polarly localized D6 PROTEIN KINASE is required for efficient auxin transport in Arabidopsis thaliana.. Development 2009; 136:627 - 36; http://dx.doi.org/10.1242/dev.028365; PMID: 19168677
- Dhonukshe P, Huang F, Galvan-Ampudia CS, Mähönen AP, Kleine-Vehn J, Xu J, et al. Plasma membrane-bound AGC3 kinases phosphorylate PIN auxin carriers at TPRXS(N/S) motifs to direct apical PIN recycling. Development 2010; 137:3245 - 55; http://dx.doi.org/10.1242/dev.052456; PMID: 20823065
- Huang F, Zago MK, Abas L, van Marion A, Galván-Ampudia CS, Offringa R. Phosphorylation of conserved PIN motifs directs Arabidopsis PIN1 polarity and auxin transport. Plant Cell 2010; 22:1129 - 42; http://dx.doi.org/10.1105/tpc.109.072678; PMID: 20407025
- Zhang J, Nodzyński T, Pencík A, Rolcík J, Friml J. PIN phosphorylation is sufficient to mediate PIN polarity and direct auxin transport. Proc Natl Acad Sci U S A 2010; 107:918 - 22; http://dx.doi.org/10.1073/pnas.0909460107; PMID: 20080776
- Ding Z, Galván-Ampudia CS, Demarsy E, Łangowski L, Kleine-Vehn J, Fan Y, et al. Light-mediated polarization of the PIN3 auxin transporter for the phototropic response in Arabidopsis. Nat Cell Biol 2011; 13:447 - 52; http://dx.doi.org/10.1038/ncb2208; PMID: 21394084
- Ganguly A, Lee SH, Cho H-T. Functional identification of the phosphorylation sites of Arabidopsis PIN-FORMED3 for its subcellular localization and biological role. Plant J 2012; In press http://dx.doi.org/10.1111/j.1365-313X.2012.05030.x; PMID: 22519832
- Dhonukshe P, Tanaka H, Goh T, Ebine K, Mähönen AP, Prasad K, et al. Generation of cell polarity in plants links endocytosis, auxin distribution and cell fate decisions. Nature 2008; 456:962 - 6; http://dx.doi.org/10.1038/nature07409; PMID: 18953331
- Sorefan K, Girin T, Liljegren SJ, Ljung K, Robles P, Galván-Ampudia CS, et al. A regulated auxin minimum is required for seed dispersal in Arabidopsis. Nature 2009; 459:583 - 6; http://dx.doi.org/10.1038/nature07875; PMID: 19478783
- Wisniewska J, Xu J, Seifertová D, Brewer PB, Ruzicka K, Blilou I, et al. Polar PIN localization directs auxin flow in plants. Science 2006; 312:883; http://dx.doi.org/10.1126/science.1121356; PMID: 16601151
- Lee OR, Kim SJ, Kim HJ, Hong JK, Ryu SB, Lee SH, et al. Phospholipase A(2) is required for PIN-FORMED protein trafficking to the plasma membrane in the Arabidopsis root. Plant Cell 2010; 22:1812 - 25; http://dx.doi.org/10.1105/tpc.110.074211; PMID: 20525850
- Aoyama T, Chua NH. A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J 1997; 11:605 - 12; http://dx.doi.org/10.1046/j.1365-313X.1997.11030605.x; PMID: 9107046
- Lee SH, Cho H-T. PINOID positively regulates auxin efflux in Arabidopsis root hair cells and tobacco cells. Plant Cell 2006; 18:1604 - 16; http://dx.doi.org/10.1105/tpc.105.035972; PMID: 16731587
- Ganguly A, Lee SH, Cho M, Lee OR, Yoo H, Cho HT. Differential auxin-transporting activities of PIN-FORMED proteins in Arabidopsis root hair cells. Plant Physiol 2010; 153:1046 - 61; http://dx.doi.org/10.1104/pp.110.156505; PMID: 20439545