Abstract
Phosphatidylinositol (4,5) bisphosphate, [PtdIns(4,5)P2], is a signaling lipid involved in many important processes in animal cells such as cytoskeleton organization, intracellular vesicular trafficking, secretion, cell motility, regulation of ion channels, and nuclear signaling pathways. In the last years PtdIns(4,5)P2 and its synthesizing enzyme, phosphatidylinositol phosphate kinase (PIPK), has been intensively studied in plant cells, revealing a key role in the control of polar tip growth. Analysis of the PIPK members from Arabidopsis thaliana, Oryza sativa and Physcomitrella patens showed that they share some regulatory features with animal PIPKs but also exert plant-specific modes of regulation. This review aims at giving an overview on the PIPK family from Arabidopsis thaliana and Physcomitrella patens. Even though their basic structure, modes of activation and physiological role is evolutionary conserved, modules responsible for plasma membrane localization are distinct for different PIPKs, depending on differences in physiological and/or developmental status of cells, such as polarized and non-polarized.
The A. thaliana PIPK family: Basis of structural properties in plant PIPKs
PtdIns(4,5)P2 is synthesized by phosphorylation in the D-5 position of the inositol ring of phosphatidylinositol-4-phosphate (PtdIns4P), by PtdIns4P 5-kinases (PIPKs).Citation1 The basic structure shared by animal, yeast and plant PIPKs consists of a dimerization domain and a highly conserved kinase domain located at the C terminus ().Citation1 In addition, most plant PIPKs contain a unique conserved domain at the N terminus, the MORN domain (Membrane Occupation and Recognition Nexus) that is characterized by repetitions of MORN motifsCitation1 and followed by a non-conserved linker region (). MORN motifs that do not contain a PIPK catalytic domain have been found in several animal and plant proteins, such as junctophilins which participate in endomembrane to plasma membrane attachment;Citation2 the MORN1 protein of Toxoplasma gondii involved in cell-division;Citation3 and the A. thaliana accumulation and replication of chloroplasts 3 protein (ARC3) involved in plastidial fission.Citation4
Figure 1. (A) Modular structure of plant type I/II B PpPIPK1. N-terminal (N-ter), MORN motifs (1–8), linker (Lin), dimerization domain (Dim), PIPK catalytic kinase domain (PIPKc) and activation loop (al). (B) Phylogenetic analysis of PIPKs. Maximum likelihood (ML) tree created with the full-length PIPKs sequences of P. patens and A. thaliana. (C) Amino acid sequence alignment of the activation loop of P. patens¸ A. thaliana, and type I and type II H. sapiens PIPKs. The asterisks indicates conserved amino acids mentioned in this review. First, two conserved positively charged amino acids (KR or KK); second, (E or A), which are involved in substrate specificity; third, (K) which is involved in plasma localization of animal type I PIPKs.
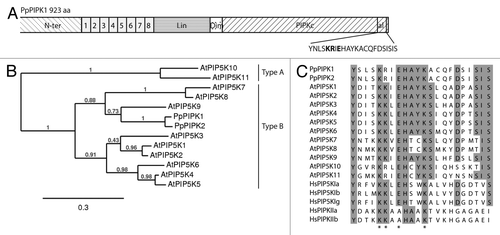
As shown in , A. thaliana contains 11 genes encoding type I/II A and B PIPKs. Subfamily A consists of two members, AtPIP5K10 and AtPIP5K11, which lack the MORN domain and exhibit a domain structure similar to human type I PIPKs, whereas the other nine isoforms (AtPIP5K1- 9) in subfamily B contain the N-terminal MORN domain.Citation1
Physcomitrella patens PIPK family
Recently we have also proceeded with the characterization of the PIPK family in the moss Physcomitrella patens. P. patens has emerged as a model system in plant biology mainly due to its high frequency of homologous recombination which allows gene targeting, thus studying gene function by direct generation of loss-of-function and point mutations on the gene of interest.Citation5,Citation6 In contrast to the 11 PIPKs encoded by the A. thaliana genome, only two isoforms are present in P. patens, PpPIPK1 and PpPIPK2, indicating a smaller gene family and less redundancy compared with flowering plants. Both PIPKs correspond to the subfamily B and no members for the A subfamily are present.Citation7 PpPIPKs amino acid sequences share 84.5% of identity, and their structure consist of eight MORN motifs at the N terminus, a linker region, and a dimerization domain followed by the catalytic kinase domain. Their kinase catalytic kinase domain is highly conserved when compared with PIPKs of other organisms, including flowering plant, human and yeast members. PpPIPK1 and PpPIPK2 belong to the clade containing AtPIP5K9 () and an identity of 55% and 57% with AtPIP5K9, respectively, among their kinase domains.
Activation modes of plant PIPKs
It has been shown that PtdIns4P is the preferred substrate in vitro for the synthesis of PtdIns(4,5)P2 by all AtPIPKs;Citation8-Citation12 this is also the case for PpPIPK1, but not for PpPIPK2, which in vitro prefers PI to produce PtdIns3P.Citation7 However, we have shown that in vivo both PpPIPK1 and PpPIPK2 catalyze the synthesis of PtdIns(4,5)P2, since the knockout lipid profile showed a reduction only in PtdIns(4,5)P2 in both pipk1 and pipk2 mutants.Citation13
Likewise to animal PIPKs, AtPIP5K1 and PpPIPK1 are activated by phosphatidic acid (PA) in vitro.Citation7,Citation14,Citation15 Whereas it has been shown that the MORN domain of AtPIP5K1 binds PtdOH and is essential for PtdOH activation,Citation14 the MORN domain does not affect PtdOH activation for PpPIPK1.Citation16 Another characteristic of type I PIPKs is their susceptibility to phosphorylation by protein kinase A, which has been shown for AtPIP5K1 and PpPIPK1.Citation7,Citation12
All PIPKs have a region within the kinase domain known as the activation loop, which contains a conserved glutamic acid residue (), responsible for the substrate specificity of animal type I PIPKs.Citation17 Similarly, the corresponding PpPIPK1E885A or AtPIP5K1E715A mutants showed an almost completely abolished activity toward PtdIns4P and PtdIns3P, but resulted in some activity with PtdIns5P, in vitro.Citation7 These results were confirmed in vivo by the fact that the phenotype of P. patens pipk1 knockout could not be completely complemented by overexpression of PpPIPK1E885A.Citation13 In addition, the dibasic amino acid pair KR in the activation loop () is also essential for the lipid kinase activity of PpPIPK1, as the mutation of KR to ND abolished activity toward PtdIns3P and PtdIns4P.Citation18
What drives PIPKs to the plasma membrane?
PIPKs are recruited to membranes but they are not integral membrane proteins. Based on the structural characteristics, one can suggest that the MORN domain is responsible for the membrane localization. For example, the MORN domain is thought to be the plasma membrane localizing module for OsPIPK1, AtPIP5K1 and AtPIP5K3.Citation14,Citation19,Citation20
Nevertheless, results obtained for AtPIP5K1, AtPIP5K2, AtPIP5K5, NtPIP5K6–1 and both PpPIPKs showed that there are other modules important for correct subcellular localization.Citation18,Citation21 In the case of PpPIPK1, we shown that the lipid kinase domain confers the plasma membrane localization through two positively charged amino acids (KR) conserved in the activation loop ().Citation18 Point mutations of the dibasic amino acid pair KR to ND in the activation loop of PpPIPK1 resulted in alternation of localization from the plasma membrane to the cytosol in P. patens protoplasts.Citation18 Importance of the kinase domain of AtPIP5K1 in plasma membrane localization was also demonstrated.Citation18 Interestingly, the kinase domain of AtPIP5K2 directs plasma membrane localization but not its apical localization in pollen tubes.Citation21 This suggests that more than one regulatory component determines apical plasma membrane localization in polarized cells. Whereas the kinase domain plays a role in recruitment of the protein to every area of the plasma membranes, the apical localization needs an additional regulatory system collaborating with the kinase domain. In contrast, AtPIP5K5 and NtPIP5K6–1 require non-conserved linker (LIM) domain for correct localization in pollen tubes, as the deletion of the N-terminal and MORN domain did not affect their apical plasma membrane localization.Citation21 The kinase domain of these two PIPKs has no function in the regulation of subcellular localization in pollen tubes.
According to the findings described previously, it is possible that protein modules responsible for plasma membrane localization are distinct in each PIPK that depends on differences in physiological and/or developmental status of cells, such as polarized and non-polarized. It is therefore necessary to compare the function of the MORN, LIM and kinase domains of every PIPKs in cells exhibiting the same stage in development. However, one should also be aware of the difficulty to compare and generalize results obtained with different experimental as well as plant systems.
The activity of PIPKs and levels of PtdIns(4,5)P2 in tip growing cells
In contrast to animal cells, cellular levels of PtdIns4P are much higher compared with PtdIns(4,5)P2 in plants, highlighting a restriction step controlling PtdIns(4,5)P2 levels by PIPKs, and thereby indicating the importance of PIPK regulation in physiological processes requiring PtdIns(4,5)P2.Citation22
In A. thaliana vegetative tissues under standard conditions, PtdIns(4,5)P2 is hard to detect which may account for the scarce information available for such tissues. Specialized plant cells such as root hairs, pollen tubes and protonemal cells of mosses (all sharing the process of cell expansion by tip growth), have been used as preferable models for studying PIPK function for several reasons. PtdIns(4,5)P2 was found to accumulate at the tip of these cellsCitation9,Citation11,Citation13,Citation23 making its detection easier by the use of fluorescence markers fused to the pleckstrin homology (PH) domain of the human PLCd1, that specifically recognizes PtdIns(4,5)P2.Citation24 These cells may have higher (and thus measurable) levels of PtdIns(4,5)P2 under standard conditions due to their high signaling activities (perceiving and transducing extracellular stimuli). Furthermore, this kind of cells can be easily followed at a single cell level. The first reports indicating the presence of PtdIns(4,5)P2 in membrane microdomains of pollen tubes or in the plasma membrane of root hair cell tips date from 1999.Citation25,Citation26 Since then it has been demonstrated that different members of the A. thaliana and moss PIPKs family play a key role in tip growth.Citation9-Citation11,Citation13,Citation19,Citation23,Citation27
From the 11 PIPK isoforms present in A. thaliana, AtPIP5K3 is specific for root hairs. Multiple lines of pip5k3 mutants exhibited reduced root hair growth, and overexpression of the gene in a wild type background resulted in deformed root hairs.Citation11 Interestingly, a AtPIP5K3 mutated version lacking the N-terminal MORN domain, but with full catalytic activity in vitro, did not complement the phenotype of pip5k3 mutants, and the overexpression of this construct resulted in deformed root hairs, meaning that AtPIP5K3 functionality in root hair development requires other factors in addition to the catalytic activity.Citation11 In pollen tubes, six different PIPK isoforms, AtPIP5K10, AtPIP5K11, AtPIP5K2, AtPIP5K4, AtPIP5K5, and AtPIP5K6 are expressed. Despite their high similarity, different roles in tip growth have been attributed to them. For example, AtPIP5K4 and AtPIP5K5 are both expressed at the apical region of the plasma membrane of pollen tubes, and pip5k4 -pip5k5 double mutants exhibited reduced pollen germination and defects in pollen tube elongation.Citation9,Citation23 Overexpression of AtPIP5K4 or AtPIP5K5 in tobacco pollen tubes cause severe growth defects, which were attributed to increased apical pectin deposition.Citation9,Citation23 AtPIP5K6 is localized at the subapical region of the plasma membrane of pollen tubes, and the suppression of AtPIP5K6 expression by RNAi resulted in impaired tip growth and inhibited clathrin-dependent endocytosis.Citation27 In contrast to AtPIP5K4, AtPIP5K5 and AtPIP5K6 which belongs to the B subfamily, AtPIP5K10 and AtPIP5K11 of the A subfamily are localized to lateral subapical plasma membrane in tobacco pollen tubes. Phenotypes observed for these latter isoforms are remarkably different from those mentioned above. Pollen tubes of pip5k10-pip5k11 double mutants, exhibited increased sensitivity to the actin polymerization inhibitor, latrunculin B, whereas overexpression of both enzymes in tobacco pollen tubes resulted in aggregation of the apical actin fine structure and a tip-swelling phenotype.Citation10 Overexpression of AtPIP5K2:EYFP, another type B isoform, resulted also in severe tip swelling of pollen tubes.Citation21 Thus the mechanisms of action of PtdIns(4,5)P2 produced by PIPKs are different, some members affect membrane trafficking and secretion, whereas others affect the actin cytoskeleton. It has been suggested that the distinct localization patterns of the enzymes may be the consequence of interactions with specific partner lipids or proteins, which recruit the enzymes to different functional microdomains.Citation10 It is clear that it is not the N-terminal MORN domain responsible for these differences in phenotypes observed between type A and B subfamilies, since overexpression of mutated isoforms lacking the MORN domain from AtPIP5K3 or AtPIP5K5 resulted in the same phenotypes as that of full-length proteins.Citation9,Citation11
In P. patens, both PIPKs are expressed in protoplasts, protonema and gametophores.Citation7 The disruption of both genes by gene targeting showed that both enzymes are involved in tip growth. Wild type moss treated with F-actin drugs resulted in a phenotype that mimicked the pipk knockout phenotype, suggesting a role of PtdIns(4,5)P2 in the cytoskeleton organization. This role was confirmed by in vivo imaging of the cytoskeleton network, which revealed that the shortened caulonemal cells in the pipk1 mutant was the result of the absence of the apicobasal gradient of cortical F-actin cables normally observed in wild type caulonemal cells.Citation13 Despite the high similarity between both proteins, a strong phenotype for pipk1 but not for pipk2 single knockout was obtained.Citation13 pipk1 knockout lines showed a dramatic growth reduction of protonema as well as of rhizoids. pipk1–2 double knockouts showed a stronger phenotype compared with pipk1; protonemal filaments exhibited an extremely compact structure and lacked the caulonemal cell type; gametophores were much shorter than the wild type with very short rhizoids and could not produce sporophytes.Citation13 Phosphoinositide analysis of pipk mutants demonstrated that a reduction of PtdIns(4,5)P2 levels was responsible for the phenotypes observed.
Future perspectives
During the last years, research about plant PIPKs has developed significantly. However, our knowledge about PIPKs and the function of its main product, PtdIns(4,5)P2 is still limited. Despite the high similarity between PIPKs and their preference for PtdIns4P, at least in vitro, other roles aside from tip growth have been described. AtPIP5K9, was shown to interact with the cytosolic invertase CINV1 to regulate sugar-mediated root cell elongation negatively.Citation28 It has recently been shown that AtPIP5K2 is involved in regulating lateral root formation and root gravity response, through regulation of PIN proteins, providing a link between the phosphatidylinositol signaling pathway and auxin response.Citation29
The function of the MORN domain still remains elusive,Citation23 but it is not excluded that it could be important for interaction with other signaling components and aid in creating signaling complexes involving PIPKs. Supporting this idea, it was shown that AtPIP5K2 interacts with small GTPases of the RabE family through the MORN domain, which may stimulate temporally or spatially localized PtdIns(4,5)P2 production at the plasma membrane.Citation30
The different functions so far ascribed to plant PIPKs points to the following open questions demanding further investigation: 1) Is a specific function related to (or dependent on) the cell-type?; 2) Is it obtained through the interaction of individual PIPKs with different interaction partners?, or 3) Is function dependent on different pools of PtdIns(4,5)P2 derived from different microdomains of the cell?
References
- Mueller-Roeber B, Pical C. Inositol phospholipid metabolism in Arabidopsis. Characterized and putative isoforms of inositol phospholipid kinase and phosphoinositide-specific phospholipase C. Plant Physiol 2002; 130:22 - 46; http://dx.doi.org/10.1104/pp.004770; PMID: 12226484
- Takeshima H, Komazaki S, Nishi M, Iino M, Kangawa K. Junctophilins: a novel family of junctional membrane complex proteins. Mol Cell 2000; 6:11 - 22; http://dx.doi.org/10.1016/S1097-2765(00)00003-4; PMID: 10949023
- Gubbels M-J, Vaishnava S, Boot N, Dubremetz JF, Striepen B. A MORN-repeat protein is a dynamic component of the Toxoplasma gondii cell division apparatus. J Cell Sci 2006; 119:2236 - 45; http://dx.doi.org/10.1242/jcs.02949; PMID: 16684814
- Maple J, Vojta L, Soll J, Møller SG. ARC3 is a stromal Z-ring accessory protein essential for plastid division. EMBO Rep 2007; 8:293 - 9; http://dx.doi.org/10.1038/sj.embor.7400902; PMID: 17304239
- Schaefer DG, Zrÿd JP. Efficient gene targeting in the moss Physcomitrella patens. Plant J 1997; 11:1195 - 206; http://dx.doi.org/10.1046/j.1365-313X.1997.11061195.x; PMID: 9225463
- Schaefer DG, Zrÿd JP. The moss Physcomitrella patens, now and then. Plant Physiol 2001; 127:1430 - 8; http://dx.doi.org/10.1104/pp.010786; PMID: 11743086
- Saavedra L, Balbi V, Dove SK, Hiwatashi Y, Mikami K, Sommarin M. Characterization of phosphatidylinositol phosphate kinases from the moss Physcomitrella patens: PpPIPK1 and PpPIPK2. Plant Cell Physiol 2009; 50:595 - 609; http://dx.doi.org/10.1093/pcp/pcp018; PMID: 19188261
- Elge S, Brearley C, Xia HJ, Kehr J, Xue HW, Mueller-Roeber B. An Arabidopsis inositol phospholipid kinase strongly expressed in procambial cells: synthesis of PtdIns(4,5)P2 and PtdIns(3,4,5)P3 in insect cells by 5-phosphorylation of precursors. Plant J 2001; 26:561 - 71; http://dx.doi.org/10.1046/j.1365-313x.2001.01051.x; PMID: 11489170
- Ischebeck T, Stenzel I, Heilmann I. Type B phosphatidylinositol-4-phosphate 5-kinases mediate Arabidopsis and Nicotiana tabacum pollen tube growth by regulating apical pectin secretion. Plant Cell 2008; 20:3312 - 30; http://dx.doi.org/10.1105/tpc.108.059568; PMID: 19060112
- Ischebeck T, Stenzel I, Hempel F, Jin X, Mosblech A, Heilmann I. Phosphatidylinositol-4,5-bisphosphate influences Nt-Rac5-mediated cell expansion in pollen tubes of Nicotiana tabacum. Plant J 2011; 65:453 - 68; http://dx.doi.org/10.1111/j.1365-313X.2010.04435.x; PMID: 21265898
- Stenzel I, Ischebeck T, König S, Hołubowska A, Sporysz M, Hause B, et al. The type B phosphatidylinositol-4-phosphate 5-kinase 3 is essential for root hair formation in Arabidopsis thaliana. Plant Cell 2008; 20:124 - 41; http://dx.doi.org/10.1105/tpc.107.052852; PMID: 18178770
- Westergren T, Dove SK, Sommarin M, Pical C. AtPIP5K1, an Arabidopsis thaliana phosphatidylinositol phosphate kinase, synthesizes PtdIns(3,4)P(2) and PtdIns(4,5)P(2) in vitro and is inhibited by phosphorylation. Biochem J 2001; 359:583 - 9; http://dx.doi.org/10.1042/0264-6021:3590583; PMID: 11672432
- Saavedra L, Balbi V, Lerche J, Mikami K, Heilmann I, Sommarin M. PIPKs are essential for rhizoid elongation and caulonemal cell development in the moss Physcomitrella patens. Plant J 2011; 67:635 - 47; http://dx.doi.org/10.1111/j.1365-313X.2011.04623.x; PMID: 21554449
- Im YJ, Davis AJ, Perera IY, Johannes E, Allen NS, Boss WF. The N-terminal membrane occupation and recognition nexus domain of Arabidopsis phosphatidylinositol phosphate kinase 1 regulates enzyme activity. J Biol Chem 2007; 282:5443 - 52; http://dx.doi.org/10.1074/jbc.M611342200; PMID: 17197438
- Perera IY, Davis AJ, Galanopoulou D, Im YJ, Boss WF. Characterization and comparative analysis of Arabidopsis phosphatidylinositol phosphate 5-kinase 10 reveals differences in Arabidopsis and human phosphatidylinositol phosphate kinases. FEBS Lett 2005; 579:3427 - 32; http://dx.doi.org/10.1016/j.febslet.2005.05.018; PMID: 15949803
- Mikami K, Saavedra L, Sommarin M. Is membrane occupation and recognition nexus domain functional in plant phosphatidylinositol phosphate kinases?. Plant Signal Behav 2010; 5:1241 - 4; http://dx.doi.org/10.4161/psb.5.10.12922; PMID: 20855959
- Kunz J, Fuelling A, Kolbe L, Anderson RA. Stereo-specific substrate recognition by phosphatidylinositol phosphate kinases is swapped by changing a single amino acid residue. J Biol Chem 2002; 277:5611 - 9; http://dx.doi.org/10.1074/jbc.M110775200; PMID: 11733501
- Mikami K, Saavedra L, Hiwatashi Y, Uji T, Hasebe M, Sommarin M. A dibasic amino acid pair conserved in the activation loop directs plasma membrane localization and is necessary for activity of plant type I/II phosphatidylinositol phosphate kinase. Plant Physiol 2010; 153:1004 - 15; http://dx.doi.org/10.1104/pp.109.152686; PMID: 20427464
- Kusano H, Testerink C, Vermeer JE, Tsuge T, Shimada H, Oka A, et al. The Arabidopsis Phosphatidylinositol Phosphate 5-Kinase PIP5K3 is a key regulator of root hair tip growth. Plant Cell 2008; 20:367 - 80; http://dx.doi.org/10.1105/tpc.107.056119; PMID: 18281506
- Ma H, Lou Y, Lin WH, Xue HW. MORN motifs in plant PIPKs are involved in the regulation of subcellular localization and phospholipid binding. Cell Res 2006; 16:466 - 78; http://dx.doi.org/10.1038/sj.cr.7310058; PMID: 16699542
- Stenzel I, Ischebeck T, Quint M, Heilmann I. Variable Regions of PI4P 5-Kinases Direct PtdIns(4,5)P(2) Toward Alternative Regulatory Functions in Tobacco Pollen Tubes. Front Plant Sci 2011; 2:114; PMID: 22639629
- Im YJ, Perera IY, Brglez I, Davis AJ, Stevenson-Paulik J, Phillippy BQ, et al. Increasing plasma membrane phosphatidylinositol(4,5)bisphosphate biosynthesis increases phosphoinositide metabolism in Nicotiana tabacum. Plant Cell 2007; 19:1603 - 16; http://dx.doi.org/10.1105/tpc.107.051367; PMID: 17496116
- Sousa E, Kost B, Malhó R. Arabidopsis phosphatidylinositol-4-monophosphate 5-kinase 4 regulates pollen tube growth and polarity by modulating membrane recycling. Plant Cell 2008; 20:3050 - 64; http://dx.doi.org/10.1105/tpc.108.058826; PMID: 19033528
- Várnai P, Balla T. Visualization of phosphoinositides that bind pleckstrin homology domains: calcium- and agonist-induced dynamic changes and relationship to myo-[3H]inositol-labeled phosphoinositide pools. J Cell Biol 1998; 143:501 - 10; http://dx.doi.org/10.1083/jcb.143.2.501; PMID: 9786958
- Braun M, Baluska F, Menzel D, Menzel D, von Witsch M. Redistribution of actin, profilin and phosphatidylinositol-4, 5-bisphosphate in growing and maturing root hairs. Planta 1999; 209:435 - 43; http://dx.doi.org/10.1007/s004250050746; PMID: 10550624
- Kost B, Lemichez E, Spielhofer P, Hong Y, Tolias K, Carpenter C, et al. Rac homologues and compartmentalized phosphatidylinositol 4, 5-bisphosphate act in a common pathway to regulate polar pollen tube growth. J Cell Biol 1999; 145:317 - 30; http://dx.doi.org/10.1083/jcb.145.2.317; PMID: 10209027
- Zhao Y, Yan A, Feijó JA, Furutani M, Takenawa T, Hwang I, et al. Phosphoinositides regulate clathrin-dependent endocytosis at the tip of pollen tubes in Arabidopsis and tobacco. Plant Cell 2010; 22:4031 - 44; http://dx.doi.org/10.1105/tpc.110.076760; PMID: 21189293
- Lou Y, Gou JY, Xue HW. PIP5K9, an Arabidopsis phosphatidylinositol monophosphate kinase, interacts with a cytosolic invertase to negatively regulate sugar-mediated root growth. Plant Cell 2007; 19:163 - 81; http://dx.doi.org/10.1105/tpc.106.045658; PMID: 17220200
- Mei Y, Jia WJ, Chu YJ, Xue HW. Arabidopsis phosphatidylinositol monophosphate 5-kinase 2 is involved in root gravitropism through regulation of polar auxin transport by affecting the cycling of PIN proteins. Cell Res 2012; 22:581 - 97; http://dx.doi.org/10.1038/cr.2011.150; PMID: 21894193
- Camacho L, Smertenko AP, Pérez-Gómez J, Hussey PJ, Moore I. Arabidopsis Rab-E GTPases exhibit a novel interaction with a plasma-membrane phosphatidylinositol-4-phosphate 5-kinase. J Cell Sci 2009; 122:4383 - 92; http://dx.doi.org/10.1242/jcs.053488; PMID: 19903693