Abstract
Mounting evidence indicate that nitric oxide (NO) acts as a signaling molecule mediating iron deficiency responses through the upregulation of the expression of iron uptake-related genes. Accordingly, NO donors such as nitrosoglutathione (GSNO) were reported to improve the fitness of plants grown under iron deficiency. Here, we showed that glutathione, a by-product of GSNO, triggered the upregulation of the expression of iron uptake- and transport-related gene and an increase of iron concentration in Arabidopsis thaliana seedlings facing iron deficiency. Furthermore, we provided evidence that under iron deficiency, NO released by GSNO did not improve the root iron concentration but impacted the content of copper. Collectively, our data highlight the complexity of interpreting data based on the use of NO donors when investigating the role of NO in iron homeostasis.
Introduction
Nitric oxide (NO) is a diatomic gas that has been shown to function as a cell-signaling molecule with diverse physiological roles in plants. Indeed, NO is involved in developmental processes, hormonal signaling and adaptive response to environmental stresses.Citation1 The mechanisms by which it exerts its effects are being discovered and include the mobilization of classical second messengers such as Ca2+ and cyclic GMP, the regulation of protein kinase activities and gene expression, the modulation of lipid signaling and the post-translational modification of numerous proteins.Citation1-Citation5
During the past 10 y, it has been recognized that NO also acts as a signaling molecule mediating iron deficiency responses.Citation6 Treatment with artificially-released NO was shown to improve the fitness of maize and tomato plants grown under iron deficiency.Citation7,Citation8 Under such conditions, the NO-treated plants displayed increased root hair development, higher chlorophyll contents and reduced interveinal chlorosis typical of iron deficiency. Further supporting these findings, NO was reported to be rapidly produced in roots of plants exposed to iron deficiency.Citation8 Once produced, it initiates an iron-starvation pathway promoting the expression of genes which products are involved in iron uptake. In Arabidopsis thaliana, these proteins include the major ferrous transporter IRT1 (Iron-Regulated Transporter 1), the root plasma membrane ferric reductase FRO2 (Ferric Reductase Oxidase 2) and the basic helix-loop-helix transcription factor FIT (Fer-like Fe deficiency-Induced Transcription factor) which positively regulates IRT1 and FRO2 expression. Accordingly, treatment of plant roots with NO donors such as nitrosoglutathione (GSNO) was reported to promote the expression of these genes or their orthologs in other plant species.Citation8 In addition, these genes were shown to be induced by NO endogenously produced in roots of A. thaliana seedlings in response to the iron deficiency caused by cadmium treatment.Citation9 The list of genes positively regulated by NO under iron deficiency also includes NAS4 (Nicotianamine Synthase 4).Citation9 The corresponding protein catalyzes the synthesis of nicotianamine, a non proteinaceous amino-acid involved in iron transport and distribution in planta.Citation10
Interestingly, although NO treatments of plants such as maize or rice were able to counteract the iron deficiency symptoms, no increase in iron content in plant organs was observed.Citation7 It has been proposed that exogenously applied NO does not improve iron uptake by roots but, instead, favors an increase in iron availability for instance through the formation of dinitrosyl iron complexes.Citation11,Citation12 These complexes, formed by the interaction between NO and ferric iron with thiols-containing ligands, could transport and deliver iron from roots to other plant organs. Furthermore, the beneficial effects of NO could be related to its ability to prevent plants from the oxidative stress caused by iron deficiency.Citation13
To further investigate the role of NO in the plant adaptive response to iron deficiency, in the present investigation we analyzed the incidence of exogenously applied NO in iron, zinc, copper and manganese accumulations in roots of A. thaliana plants exposed to iron deficiency.
Results
A. thaliana seedlings grown for 4 weeks in hydroponic conditions with 50 µM of iron were transferred for 96 h in the same medium in which the concentration of iron was reduced to 5 µM. Forty eight hours after the transfer, plant roots were exposed to 50 µM of the NO donors GSNO or diethylamine-NONOate (DEANO). Then, after 48 h of incubation with the NO donors, the root metal concentrations were examined by induced coupled plasma-optical emission spectrometry (ICP-OES). To check whether the NO donor treatments impact the expression of iron-uptake/transport genes in our conditions, the accumulation of IRT1, FRO2 and NAS4 in roots were analyzed 24 h after the supplementation of the medium with these compounds.
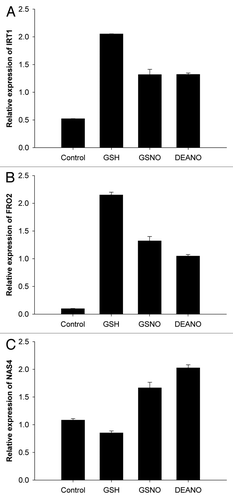
Compared with the control plants, GSNO triggered a 2.5 and 13-fold increase of IRT1 and FRO2 transcript accumulation, respectively (). These data fit well with those of Graziano and LamattinaCitation8 who reported that GSNO mediates the expression of the orthologs of the A. thaliana IRT1 and FRO2 genes in tomato plants under iron deprivation. Because GSNO releases both glutathione (GSH) and NO, the putative effect of GSH was also assessed (). Surprisingly, GSH induced the strongest upregulation of IRT1 and FRO2 expression. Indeed, compared with GSNO treatment, the levels of IRT1 and FRO2 mRNA were increased by 1.6 fold. To confirm that the effect of GSNO was at least partly caused by NO released by the donor, plants were also exposed to the NO donor DEANO. Whereas diethylamine alone did not impact the accumulation of the transcripts of interest (data not shown), DEANO promoted the expression of both genes by 2.6 and 11.5 fold as compared with control plants, respectively. Collectively, these data indicate that NO as well as GSH treatments amplify the accumulation of IRT1 and FRO2 transcripts in roots of A. thaliana seedlings grown in iron deficient condition. Interestingly, different data were obtained regarding NAS4 transcripts (). Indeed, whereas a 1.5 and 1.8 fold increase of NAS4 mRNA accumulation occurred in response to GSNO and DEANO, respectively, GSH treatment slightly reduced their levels. Therefore, the regulation of the expression of NAS4 might differ from those of IRT1 and FRO2.
Figure 1. Incidence of GSNO, DEANO and GSH on the accumulation of IRT1, FRO2 and NAS4 transcripts in roots of plants exposed to iron deficiency. (A) IRT1 transcript accumulation, (B) FRO2 transcript accumulation, (C) AtNAS4 transcript accumulation in roots. Plants were cultivated 4 weeks in iron-sufficient conditions (Fe 50 µM) and then transferred 48 h in iron deficient (Fe 5 µM) conditions. Then, plants were treated with GSNO (50 µM), GSH (50 µM) or DEANO (50 µM) for 24 h. Transcript accumulation of IRT1, FRO2 and NAS4 in roots were measured by qRT-PCR. Data are expressed as mean normalized expression relative to the appropriate housekeeping gene AtYLS8 (arbitrary units) and are the means of triplicates (± SE).
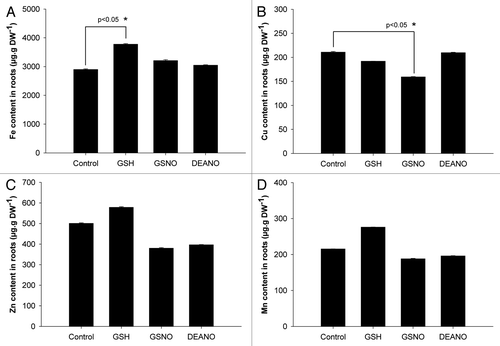
Concerning the concentration of iron, no differences were observed between control plants and plants exposed to GSNO or DEANO (). In contrast, compared with WT plants, iron concentration was significantly increased by 1.3 fold in response to GSH. Regarding copper (), while its concentration was unchanged in GSH- and DEANO-treated plants compared with control plants, differences were observed in response to GSNO treatment. Indeed, compared with WT plants, the concentration of copper was significantly decreased by 25%. In the case of zinc and manganese, no significant differences were observed between control plants and plants exposed to GSH, GSNO or DEANO (). Taken together, these data indicate that GSNO impacted the concentration of copper. However, both NO donors did not improve the iron concentration of plants grown under iron deficiency, thus confirming Graziano et al.Citation7 previous observation in maize. Furthermore, GSH itself affected the concentration of iron.
Figure 2. Impact of GSNO, DEANO and GSH on the concentrations of iron, manganese, zinc and copper in roots of plants exposed to iron deficiency. (A) Iron concentration, (B) copper concentration, (C) zinc concentration, (D) manganese concentration. Plants were cultivated 4 weeks in iron-sufficient conditions (Fe 50 µM) and then transferred 48 h in iron deficient (Fe 5 µM) conditions. Then, plants were treated with GSNO (50 µM), GSH (50 µM) or DEANO (50 µM) for 48 h. The concentration of metals was determined in the roots by ICP-OES. Each value represents the means ± SD of nine measurements (three replicates per experiment performed three times). Stars represent significant differences from control according to ANOVA on ranks followed by a comparison vs. control (p < 0.05).
Discussion
The present investigation confirmed previous data showing that NO released by NO donors promotes the expression of IRT1 and FRO2 in plants suffering from iron deficiency without favoring an increase of iron uptake. It also confirms our recent study highlighting a role for NO in the regulation of the expression of NAS4.Citation9 The observation that GSH, a by-product of GSNO, also influenced the plant response to iron deficiency raises the question of the role of GSH in iron deficiency as well as its interplay with NO. It has been previously reported that iron deficiency enhances the level of GSH.Citation14 According to Ramirez et al.,Citation11 GSH might act as a thiol-containing ligand in the formation of dinitrosyl iron complexes and, consequently, might contribute to the increase of iron availability in plant tissues. Furthermore, it seems plausible that the feeding of plants with GSH might increase the intracellular pool of GSNO which constitutes a stable and mobile endogenous reservoir of NO. The rise in GSNO content could promote the NO-dependent mechanisms contributing to the iron deficiency responses such as the upregulation of IRT1 and FRO2 transcript expression. Besides these aspects, our work also highlights the importance of GSH as a control when assessing the effects of GSNO on plant biological processes.
Others important findings of the present investigation concern the influence of the NO donors on the accumulation of other metals. Indeed, our data indicate that GSNO also impacted the concentration of copper. This observation is not surprising as the ion homeostasis networks in plants seem closely linked.Citation15 Notably, iron, manganese and zinc are all taken up by IRT1 and FRO2 is also able to reduce Cu.Citation16 Therefore, copper, iron, zinc and manganese might affect each other’s uptake.
Finally, our data showed that the plant response to NO might differ according to the NO donor. Indeed, GSNO and not DEANO triggered a decrease in the concentration of copper. Beside the fact that GSNO releases both NO and GSH, GSNO mainly releases nitrosonium cation (NO+) while DEANO releases NO in its radical form (NO•). Furthermore, the kinetic of NO release differs between both NO donors. Indeed, the release of NO from DEANO occurs within seconds in aqueous solutionCitation17 whereas the release form GSNO is slower.Citation18 Although speculative, we assume that these parameters might influence the action of NO. Here too, this discussion highlights the complexity of interpreting data based on the use of NO donors.
Materials and Methods
Plant material and growth conditions
Experiments were performed using A. thaliana Columbia ecotype (Col-0) wild-type. Seeds were surface sterilized by immersion in a solution containing 1.2% bleach and 50% EtOH for 5 min and placed for 3 d at 4°C in 0.15% agar. Then, they were sown in seed holders filled with 0.75% agar and put in a cover placed above the nutrient solution container (Araponics). The medium (0.25 mM Ca(NO3)2, 0.5 mM KNO3, 1 mM KH2PO4, 1 mM MgSO4, 50 µM H3BO3, 19 µM MnCl2, 10 µM ZnCl2, 1 µM CuSO4, 0.02 µM Na2MoO4, 50 µM Fe-Na-EDTA) was changed once a week. Plants were allowed to grow for 4 weeks in the following conditions: 10 h day (200 µE.m−2.s−1 light intensity, 20°C) / 14 h night (18°C) with 70% relative humidity. After 4 weeks of culture, roots were washed 3 times with the nutritive solution containing 5 µM Fe-EDTA instead of 50 µM. Then, plants were grown for 96 h on the hydroponic medium with 5 µM Fe- EDTA.
Plant treatments
Forty eight hours after their transfer into iron-deficient conditions, plants were treated with 50 µM of GSNO, GSH or DEANO. GSNO and GSH were dissolved in water, DEANO was prepared as previously described.Citation17 As a control for DEANO effects, plants were treated with 50 µM diethylamine prepared in water.
Determination of metal concentration
Roots were collected 48 h after the addition of the NO donors or GSH into the culture medium. The concentration of metals was determined in the roots by induced coupled plasma-optical emission spectrometry (ICP-OES) as previously reported.Citation9
qRT-PCR analyses
Root tissues were collected after the treatments and immediately frozen in liquid nitrogen before total RNA extraction using Trizol reagent according to the manufacturer’s instructions (Invitrogen). A DNase (Promega) treatment was performed on 5 µg of total RNA to prevent genomic DNA contamination. RNA samples were subsequently used for reverse transcription (ImpromIITM Reverse Transcriptase kit, Promega) with anchored oligo(dT15) (Promega) and 0.4 mM deoxynucleotide triphosphates. The resulting cDNAs were subjected to a 2-fold dilution with water and one µl of each cDNA sample was assayed by qPCR (7900HT, Applied Biosystems®). Expression levels were calculated relatively to the housekeeping gene AtYLS8 (At5g08290) using the relative standard curve method. For each sample, target quantity of the gene of interest was determined by interpolating the value from the standard curve made from a cDNA pool which enables to take into consideration the efficiency of amplification. The value was then divided by the target quantity of the housekeeping gene.
Primer sequences were as follows: AtNAS4-fw: 5′- AGAAGGTTAAGGTGGTCGAGC-3′, AtNAS4-rev: 5′-ACAGTTACACGCGAGATCCGA-3′; IRT1-fw: 5′-CGGTTGGACTTCTAAATGC-3′, IRT1-rev: 5′-CGATAATCGACATTCCACCG-3′; FRO2-fw: 5′-CGATCGTTTCCTTCGGTTTC-3′, FRO2-rev: 5′-AATCCGAGCAGCGAGCAA-3′; AtYLS8-fw: 5′-GTCATTTATCTGGTGGACATCACT-3′, AtYLS8-rev: 5′-CCGAAACAGTAAGCAAAAAC-3′.
Chemicals
All chemicals were purchased from Sigma-Aldrich.
Statistical analysis
In order to detect significant differences between the different treatments, ANOVA on ranks were performed with the statistical program SigmaPlot release 11.0 (Systat Software) followed by a comparison vs. control with the Dunn’s Method (* p < 0.05).
| Abbreviations: | ||
| DEANO | = | diethylamine-NONOate |
| FIT | = | Fer-like Fe deficiency-Induced Transcription factor |
| FRO2 | = | Ferric Reductase Oxidase 2 |
| GSNO | = | nitrosoglutathione |
| ICP-OES | = | coupled plasma-optical emission spectrometry |
| IRT1 | = | Iron-Regulated Transporter 1 |
| NAS4 | = | Nicotianamine Synthase 4 |
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This was supported by the Agence Nationale de la Recherche (program BLAN07–2_184783 PIANO) and the University of Burgundy (FABER project to A. B-B).
References
- Besson-Bard A, Pugin A, Wendehenne D. New insights into nitric oxide signaling in plants. Annu Rev Plant Biol 2008; 59:21 - 39; http://dx.doi.org/10.1146/annurev.arplant.59.032607.092830; PMID: 18031216
- Astier J, Kulik A, Koen E, Besson-Bard A, Bourque S, Jeandroz S, et al. Protein S-nitrosylation: What’s going on in plants?. Free Radic Biol Med 2012; In press http://dx.doi.org/10.1016/j.freeradbiomed.2012.06.032; PMID: 22750205
- Baudouin E. The language of nitric oxide signalling. Plant Biol (Stuttg) 2011; 13:233 - 42; http://dx.doi.org/10.1111/j.1438-8677.2010.00403.x; PMID: 21309969
- Gaupels F, Kuruthukulangarakoola GT, Durner J. Upstream and downstream signals of nitric oxide in pathogen defence. Curr Opin Plant Biol 2011; 14:707 - 14; http://dx.doi.org/10.1016/j.pbi.2011.07.005; PMID: 21816662
- Leitner M, Vandelle E, Gaupels F, Bellin D, Delledonne M. NO signals in the haze: nitric oxide signalling in plant defence. Curr Opin Plant Biol 2009; 12:451 - 8; http://dx.doi.org/10.1016/j.pbi.2009.05.012; PMID: 19608448
- Graziano M, Lamattina L. Nitric oxide and iron in plants: an emerging and converging story. Trends Plant Sci 2005; 10:4 - 8; http://dx.doi.org/10.1016/j.tplants.2004.12.004; PMID: 15642517
- Graziano M, Beligni MV, Lamattina L. Nitric oxide improves internal iron availability in plants. Plant Physiol 2002; 130:1852 - 9; http://dx.doi.org/10.1104/pp.009076; PMID: 12481068
- Graziano M, Lamattina L. Nitric oxide accumulation is required for molecular and physiological responses to iron deficiency in tomato roots. Plant J 2007; 52:949 - 60; http://dx.doi.org/10.1111/j.1365-313X.2007.03283.x; PMID: 17892445
- Besson-Bard A, Gravot A, Richaud P, Auroy P, Duc C, Gaymard F, et al. Nitric oxide contributes to cadmium toxicity in Arabidopsis by promoting cadmium accumulation in roots and by up-regulating genes related to iron uptake. Plant Physiol 2009; 149:1302 - 15; http://dx.doi.org/10.1104/pp.108.133348; PMID: 19168643
- Lee S, Jeon US, Lee SJ, Kim YK, Persson DP, Husted S, et al. Iron fortification of rice seeds through activation of the nicotianamine synthase gene. Proc Natl Acad Sci U S A 2009; 106:22014 - 9; http://dx.doi.org/10.1073/pnas.0910950106; PMID: 20080803
- Ramirez L, Zabaleta EJ, Lamattina L. Nitric oxide and frataxin: two players contributing to maintain cellular iron homeostasis. Ann Bot 2010; 105:801 - 10; http://dx.doi.org/10.1093/aob/mcp147; PMID: 19556267
- Jasid S, Simontacchi M, Puntarulo S. Exposure to nitric oxide protects against oxidative damage but increases the labile iron pool in sorghum embryonic axes. J Exp Bot 2008; 59:3953 - 62; http://dx.doi.org/10.1093/jxb/ern235; PMID: 18832188
- Sun B, Jing Y, Chen K, Song L, Chen F, Zhang L. Protective effect of nitric oxide on iron deficiency-induced oxidative stress in maize (Zea mays). J Plant Physiol 2007; 164:536 - 43; http://dx.doi.org/10.1016/j.jplph.2006.02.011; PMID: 16690167
- Zaharieva TB, Abadía J. Iron deficiency enhances the levels of ascorbate, glutathione, and related enzymes in sugar beet roots. Protoplasma 2003; 221:269 - 75; PMID: 12802634
- Lahner B, Gong J, Mahmoudian M, Smith EL, Abid KB, Rogers EE, et al. Genomic scale profiling of nutrient and trace elements in Arabidopsis thaliana.. Nat Biotechnol 2003; 21:1215 - 21; http://dx.doi.org/10.1038/nbt865; PMID: 12949535
- Pilon M, Cohu CM, Ravet K, Abdel-Ghany SE, Gaymard F. Essential transition metal homeostasis in plants. Curr Opin Plant Biol 2009; 12:347 - 57; http://dx.doi.org/10.1016/j.pbi.2009.04.011; PMID: 19481497
- Besson-Bard A, Griveau S, Bedioui F, Wendehenne D. Real-time electrochemical detection of extracellular nitric oxide in tobacco cells exposed to cryptogein, an elicitor of defence responses. J Exp Bot 2008; 59:3407 - 14; http://dx.doi.org/10.1093/jxb/ern189; PMID: 18653691
- Singh SP, Wishnok JS, Keshive M, Deen WM, Tannenbaum SR. The chemistry of the S-nitrosoglutathione/glutathione system. Proc Natl Acad Sci U S A 1996; 93:14428 - 33; http://dx.doi.org/10.1073/pnas.93.25.14428; PMID: 8962068