Abstract
Transcriptionally active chromosome (TAC) is a fraction of protein/DNA complexes with RNA polymerase activity in the plastid. The function of most TAC proteins is not well known. We isolated a mutant gene encoding a plastid TAC component, pTAC14, and performed functional analysis of plastid gene expression and chloroplast development in Arabidopsis. We found that knockout of pTAC14 led to the blockage of thylakoid formation in the initial process of chloroplast development. Furthermore, the transcript levels of plastid-encoded polymerase (PEP)-dependent genes were downregulated in ptac14, suggesting that PEP activity was decreased in the mutant. On the basis of these results, we briefly review the available evidence and highlight the interaction between pTAC14 and pTAC12 that could help us understand the regulatory role of pTAC14 in chloroplast development and plastid gene expression.
Arabidopsis pTAC14 Proteins
The chloroplast is a semiautonomous organelle that requires the coordination of nuclear and chloroplast genes.Citation1 The expression of plastid-encoded genes is performed by at least 2 RNA polymerases:Citation2,Citation3 one is a plastid-encoded polymerase (PEP) and the other is a nuclear-encoded polymerase (NEP). In general, PEP is involved in the expression of photosynthesis-related genes, and PEP-dependent genes are transcribed late in chloroplast development.Citation2,Citation4 Although the core subunits of the PEP complex are plastid encoded, participation of nuclear-encoded factors is required for the transcription activity of PEP. Plastid sigma factors are considered to play a key role in PEP regulation.Citation5 There are 6 sigma factors (SIG1–SIG6) in Arabidopsis. Moreover, the transcriptional activity of PEP also requires other nuclear-encoded factors. Several reports indicate that PEP is responsible for the transcription of plastid genes involved in photosynthesis.Citation6-Citation9 PEP-associated proteins have been isolated through purification of the PEP complex components, and they are referred to as plastid transcriptionally active chromosome (pTAC) proteins. However, the function of pTACs remains unclear.Citation6 pTAC14 is one component of the PEP complex. We found that pTAC14 is a chloroplast -located protein, and it is highly conserved and specific in plants. It contains 2 domains: the truncated SET domain and a Rubisco LSMT substrate-binding domain.Citation10 Both of them have putative methyltransferase activity targeted to nonhistone proteins.Citation11-Citation13 By genetic analysis, genomic complementation experiments, and albino phenotype analysis of the knockout mutant, we found that pTAC14 is important for early chloroplast development; furthermore, a null mutation of pTAC14 leads to the albino phenotype and failed formation of the thylakoid membrane.
pTAC14 is Important for the PEP Activity
TACs are important components of a complex that is required for PEP activity in plastids.Citation6,Citation14,Citation15 By using real-time RT-PCR and northern blot hybridization, we observed decreased PEP activity in a ptac14 mutant and hypothesized that pTAC14 is important for the maintenance of PEP activity. Steiner et al.Citation16 have provided a subunit catalog of the basic PEP complex. The deletion or knockdown lines for these components, including ptac2,Citation6 ptac6,Citation6 ptac12,Citation6 atmurE,Citation14 fsd2 and fsd,Citation15 trxZ,Citation17 and fln1,Citation16,Citation17 displayed albinism and/or seedling lethality and decreased PEP-dependent chloroplast gene expression. These results were in accordance with our conclusion. Furthermore, transcripts transcribed by SIG6 cannot be detected in ptac14. In PEP-deficient mutants, transcripts initiated by NEP would cover the entire plastome when no functional sigma factor is available for PEP. This finding suggested that pTAC14 is involved in SIG6 function. However, it is equally possible that the low PEP activity in ptac14 was insufficient to initiate transcription of PEP-dependent mRNAs. Conversely, we found that NEP-dependent transcription was upregulated in the ptac14 mutant. This finding was in agreement with other reported PEP-deficient mutants such as ys1,Citation7 clb19,Citation8 and dg1.Citation9 Although the molecular mechanism of the upregulation is not clear, it is generally thought that a feedback mechanism mediated by tRNAPGlu in the mutant explains the high NEP activity.Citation18 Therefore, it is unlikely that the knockout lethal phenotype is correlated with increased levels of NEP-dependent genes.
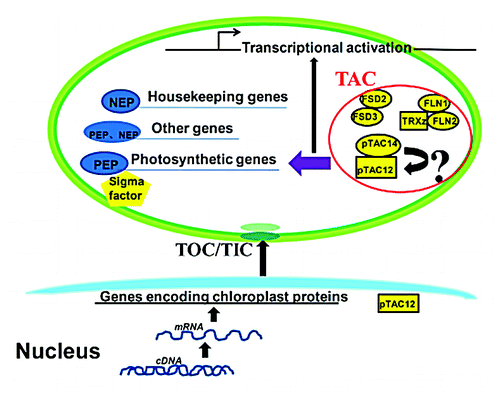
In order to better illustrate the molecular mechanism of pTAC14 function, we obtained further insight into the relationship of pTAC14 with other known TAC components and identified an interacting protein, pTAC12, in chloroplasts. Thus far, no biochemical data are available to demonstrate physiological roles of the interaction in plant development. However, the albino phenotype of the knockout lines for the 2 genes, combined with the decreased PEP-dependent chloroplast gene expression pattern suggests a functional interaction between pTAC14 and pTAC12 in TACs. These 2 proteins likely work together to perform essential roles in regulating PEP activity,Citation6,Citation10 and this is important for the chloroplast development.
Conclusions and Perspectives
Despite the isolation of several TACs and characterization of some individual members, the functional interactions between various components remain unclear. Previous reports identified pTAC12 as HEMERA, which was localized in both the chloroplast and nucleus.Citation19 However, its function in the chloroplast is still ambiguous. Our results demonstrated that pTAC14 is directly associated with pTAC12 in the chloroplast, which is a key component of the phytochrome signaling pathway that regulates PEP activities and plastid gene expression. This provides a new direction and a new perspective for the research of pTAC12 function in the chloroplast. On the other hand, it is important to analyze pTAC14 activity and to address the physiological significance of the interaction of pTAC14 and pTAC12/HEMERA proteins in plant cells. As both Rubisco LSMT substrate-binding domain and a truncated SET domain in pTAC14 are predicted to confer the methyltransferase activity, we propose that pTAC14 may regulate the function of pTAC12 and other TAC components through methylation, thereby affecting PEP activity in the chloroplast (). We suggest that site-specific mutation experiments within key residues of the SET-domain, coupled with genomic complementation analysis, should be performed. Following this approach, we can then indirectly confirm pTAC14 methylation activity if the transgenic complementary plants recover the SET-domain function. In addition, global methylation studies in Arabidopsis chloroplasts should be a goal of future research. Although we found interaction between pTAC14 and pTAC12, comprehensive analyses, including mapping of the regions responsible for the interaction, and determination of the environmental factors that affect PEP activity and plastid gene expression, will help to resolve outstanding issues. These approaches will shed more light on the molecular mechanisms by which TAC components regulate plastid gene expression.
Acknowledgments
This investigation is supported by grants from the National Basic Research Program of China (2009CB118504), Shanghai Municipal Natural Science Foundation (10ZR1421800), and the Leading Academic Discipline Project of Shanghai Municipal Education Commission (J50401); it is funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
References
- Mullet JE. Chloroplast development and gene expression. Annu Rev Plant Physiol Plant Mol Biol 1988; 39:475 - 502; http://dx.doi.org/10.1146/annurev.pp.39.060188.002355
- Lopez-Juez E, Pyke KA. Plastids unleashed: their development and their integration in plant development. Int J Dev Biol 2005; 49:557 - 77; http://dx.doi.org/10.1387/ijdb.051997el; PMID: 16096965
- Pyke K. Plastid biogenesis and differentiation. Curr Genet 2007; 19:1 - 28; http://dx.doi.org/10.1007/4735_2007_0226
- Hajdukiewicz PTJ, Allison LA, Maliga P. The two RNA polymerases encoded by the nuclear and the plastid compartments transcribe distinct groups of genes in tobacco plastids. EMBO J 1997; 16:4041 - 8; http://dx.doi.org/10.1093/emboj/16.13.4041; PMID: 9233813
- Schweer J, Loschelder H, Link G. A promoter switch that can rescue a plant sigma factor mutant. FEBS Lett 2006; 580:6617 - 22; http://dx.doi.org/10.1016/j.febslet.2006.11.010; PMID: 17118361
- Pfalz J, Liere K, Kandlbinder A, Dietz KJ, Oelmüller R. pTAC2, -6, and -12 are components of the transcriptionally active plastid chromosome that are required for plastid gene expression. Plant Cell 2006; 18:176 - 97; http://dx.doi.org/10.1105/tpc.105.036392; PMID: 16326926
- Zhou WB, Cheng YX, Yap A, Chateigner-Boutin AL, Delannoy E, Hammani K, et al. The Arabidopsis gene YS1 encoding a DYW protein is required for editing of rpoB transcripts and the rapid development of chloroplasts during early growth. Plant J 2009; 58:82 - 96; http://dx.doi.org/10.1111/j.1365-313X.2008.03766.x; PMID: 19054358
- Chateigner-Boutin AL, Ramos-Vega M, Guevara-García A, Andrés C, de la Luz Gutiérrez-Nava M, Cantero A, et al. CLB19, a pentatricopeptide repeat protein required for editing of rpoA and clpP chloroplast transcripts. Plant J 2008; 56:590 - 602; http://dx.doi.org/10.1111/j.1365-313X.2008.03634.x; PMID: 18657233
- Chi W, Ma JF, Zhang DY, Guo JK, Chen F, Lu CM, et al. The pentratricopeptide repeat protein DELAYED GREENING1 is involved in the regulation of early chloroplast development and chloroplast gene expression in Arabidopsis. Plant Physiol 2008; 147:573 - 84; http://dx.doi.org/10.1104/pp.108.116194; PMID: 18400937
- Gao ZP, Yu QB, Zhao TT, Ma Q, Chen GX, Yang ZN. A functional component of the transcriptionally active chromosome complex, Arabidopsis pTAC14, interacts with pTAC12/HEMERA and regulates plastid gene expression. Plant Physiol 2011; 157:1733 - 45; http://dx.doi.org/10.1104/pp.111.184762; PMID: 22010110
- Ying ZT, Mulligan RM, Janney N, Houtz RL. Rubisco small and large subunit N-methyltransferases. Bi- and mono-functional methyltransferases that methylate the small and large subunits of Rubisco. J Biol Chem 1999; 274:36750 - 6; http://dx.doi.org/10.1074/jbc.274.51.36750; PMID: 10593982
- Couture JF, Hauk G, Thompson MJ, Blackburn GM, Trievel RC. Catalytic roles for carbon-oxygen hydrogen bonding in SET domain lysine methyltransferases. J Biol Chem 2006; 281:19280 - 7; http://dx.doi.org/10.1074/jbc.M602257200; PMID: 16682405
- Ng DW, Wang T, Chandrasekharan MB, Aramayo R, Kertbundit S and Hall TC. Plant SET domain-containing proteins: structure, function and regulation. Biochim Biophys Acta 2007; 1769:316-29; PMID: 17512990; DOI: 10.1016/j.bbaexp.2007.04.003.
- Garcia M, Myouga F, Takechi K, Sato H, Nabeshima K, Nagata N, et al. An Arabidopsis homolog of the bacterial peptidoglycan synthesis enzyme MurE has an essential role in chloroplast development. Plant J 2008; 53:924 - 34; http://dx.doi.org/10.1111/j.1365-313X.2007.03379.x; PMID: 18036201
- Myouga F, Hosoda C, Umezawa T, Iizumi H, Kuromori T, Motohashi R, et al. A heterocomplex of iron superoxide dismutases defends chloroplast nucleoids against oxidative stress and is essential for chloroplast development in Arabidopsis. Plant Cell 2008; 20:3148 - 62; http://dx.doi.org/10.1105/tpc.108.061341; PMID: 18996978
- Steiner S, Schröter Y, Pfalz J, Pfannschmidt T. Identification of essential subunits in the plastid-encoded RNA polymerase complex reveals building blocks for proper plastid development. Plant Physiol 2011; 157:1043 - 55; http://dx.doi.org/10.1104/pp.111.184515; PMID: 21949211
- Arsova B, Hoja U, Wimmelbacher M, Greiner E, Ustün S, Melzer M, et al. Plastidial thioredoxin z interacts with two fructokinase-like proteins in a thiol-dependent manner: evidence for an essential role in chloroplast development in Arabidopsis and Nicotiana benthamiana. Plant Cell 2010; 22:1498 - 515; http://dx.doi.org/10.1105/tpc.109.071001; PMID: 20511297
- Hanaoka M, Kanamaru K, Fujiwara M, Takahashi H, Tanaka K. Glutamyl-tRNA mediates a switch in RNA polymerase use during chloroplast biogenesis. EMBO Rep 2005; 6:545 - 50; http://dx.doi.org/10.1038/sj.embor.7400411; PMID: 15877080
- Chen M, Galvão RM, Li M, Burger B, Bugea J, Bolado J, et al. Arabidopsis HEMERA/pTAC12 initiates photomorphogenesis by phytochromes. Cell 2010; 141:1230 - 40; http://dx.doi.org/10.1016/j.cell.2010.05.007; PMID: 20603003