Abstract
In plant cells, diverse environmental changes often induce transient elevation in the intracellular calcium concentrations, which are involved in signaling pathways leading to the respective cellular reactions. Therefore, these calcium elevations need to be deciphered into specific downstream responses. Calmodulin-like-proteins (CMLs) are calcium-sensing proteins present only in higher plants. They are involved in signaling processes induced by both abiotic as well as biotic stress factors. However, the role of CMLs in the interaction of plants with herbivorous insects is almost unknown. Here we show that in Arabidopsis thaliana a number of CMLs genes (CML9, 11,12,16,17 and 23) are upregulated due to treatments with oral secretion of larvae of the herbivorous insect Spodoptera littoralis. We identified that these genes belong to two groups that respond with different kinetics to the treatment with oral secretion. Our data indicate that signaling networks involving multiple CMLs very likely have important functions in plant defense against insect herbivores, in addition to their involvement in many other stress-induced processes in plants.
Throughout their life, plants are challenged by various abiotic and biotic changes in their environment. Any appropriate reaction to such environmental variations needs the recognition of the respective information, followed by downstream intracellular signaling leading to a specific response. Most of the time, upon perception of stress signals, a transient increase in the cytosolic calcium (Ca2+) concentration can be observed in plant cells.Citation1,Citation2 However, Ca2+ elevations due to stress signals are rather ubiquitous, general responses. One of the determinants of specificity is Ca2+ signature, specific to a stimulus, characterized by its duration, amplitude, frequency, and location; the other is presence of Ca2+ sensor proteins that contribute to the induction of specific physiological response.Citation3 In plants, Ca2+ sensor proteins are classified as sensor responders, (e.g., calmodulin-dependent protein kinases, CDPKs) and sensor relay proteins. The latter proteins only undergo conformational changes upon Ca2+ binding and subsequently interact with target proteins. Calmodulin-like proteins (CMLs) are sensor relay proteins which are unique to plants with 50 members in Arabidopsis thaliana.Citation4 They possess 2 to 6 predicted Ca2+-binding EF hand motifs. With calmodulins, CMLs share at least 15% identity on the amino acid level.Citation4,Citation5 CMLs are involved in stress perception and plant development. For example, CML24 is known to cause alterations in flowering time, abscisic acid (ABA) level and ion stress;Citation6,Citation7 CML37, CML38 and CML39 transcripts are regulated by abiotic stress (salt and drought), phytohormones (jasmonate and ABA), and biotic stress (phytopathogenic Pseudomonas syringae).Citation8 CML8 is induced by salicylic acid (SA) and salt stress.Citation9 CML9 alters plant responses to ABA and abiotic stress and CML9 gene is induced by infection with P. syringae, flg22 elicitor, and SA.Citation10,Citation11 Only recently it was demonstrated that CML42 represents both a negative regulator of insect herbivory-induced defense, drought induced ABA levels and a positive regulator of UV stress.Citation12 Moreover, loss of CML42 function leads to aberrant trichomes with increased branching.Citation13
We demonstrated that cellular calcium (Ca2+) elevation is an early event in the interaction between S. littoralis and A. thaliana. Up to now, among the numerous CMLs only one (CML42) has been described to be involved in plant response to insect herbivory.Citation12 In order to identify new herbivory-related targets in the CML gene family, we investigated the transcript level of various CML genes in A. thaliana upon treatment with oral secretions (OS) from larvae of the generalist herbivore S. littoralis. Preliminary microarray analyses using an Affymetrix array revealed that besides the strongly induced CML42 (At4g20780),Citation12 other CMLs were regulated by S. littoralis OS application as well. Here, using quantitative real time PCR, we further confirmed and explored the regulation of CMLs in more detail.
Therefore, A. thaliana leaves were wounded and treated with either water (control) or S. littoralis OS and CMLs expression was analyzed in these samples. We found that in the 50 member gene family of CMLs, mainly CML9, 11, 12, 16, 17, and 23 are upregulated by elicitors present in the OS of S. littoralis. A further time course experiment of gene expression revealed that expressions could be classified into two main groups: (1) Early and transiently expressed CMLs- CML11, 12, and 16; and (2) late and sustained expressed CMLs- CML9, 17 and 23.
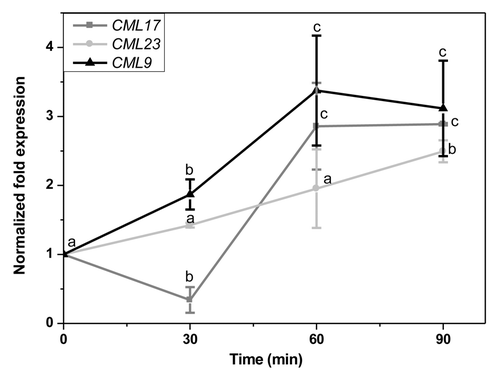
Group 1 genes (CML11, 12, and 16) showed their highest upregulation 30 min after treatment. Their level of expression decreased at later time points (). In this set, CML12/TCH3 was the most highly upregulated gene with expression levels reaching 9-fold of the control within 30 min. This level of expression decreased with time to 5-fold. CML11 and 16 responded to OS treatment with lower expression levels. They also showed maximal upregulation at 30 min but reached basal levels after 60 min. CML12/TCH3, encodes a unique calmodulin-like protein, with 6 putative Ca2+ binding EF hands as opposed to 4 or fewer EF hands in other CMLs. It is rapidly induced by mechanical stimulation, ethylene, auxin, cold, and extracellular calcium.Citation14-Citation18 Plants respond to two concomitant stimuli of herbivory: mechanical wounding and recognition of elicitors in OS.The sustained expression of CML12 could be due to the fact that it is also upregulated by mechanical wounding alone and OS might act to amplify the wound-induced signal. The recently described CML42 also belongs to this group 1.Citation12 CMLs might also be regulated by jasmonates, apart from direct herbivory signals, due to the jasmonate burst in plants upon S. littoralis OS treatment.Citation12 Stimuli-induced CMLs gene expressionCitation5 upon various stress treatment has revealed the group 1 genes, CML12 and CML16 are not regulated by methyl jasmonate (MeJA). In fact, CML16 expression was downregulated by MeJA.Citation5 The group 2 genes include CML9, 17, and 23. They are characterized by a sustained expression upon treatment with S. littoralis OS. They reached their maximal level of expression at 60 or 90 min after treatment (). Within this group, CMl17 is unique because it was initially downregulated and upregulated only at 60 and 90 min, whereas CML9 and 23 showed a steady increase over time. In this group both, CML23 and 17 are MeJA-induced genes. This further point to the fact that sustained expression in this group might be due to combined action of both oral secretions and JA burst. CML9 however is an exception and is a MeJA-repressed gene.Citation5 CML23 and CML24 are known to be potential calcium sensors that have partially overlapping function and regulate nitric oxide accumulation and transition to flowering.Citation19 However, the overall fold-change of expression in group 2 was lesser than in group 1. We thus identified 6 new target genes in A. thaliana which respond to elicitors in S. littoralis OS and might be involved in plant defense.
Figure 1.CML11, CML12, and CML16 transcript levels in leaves of A. thaliana 30, 60 and 90 min after treatment with Spodoptera littoralis oral secretion (OS). Leaves were elicited by pattern wheel wounding and subsequently treating the wound with 20 µL water or 1:1 diluted OS per leaf. Transcript abundance in leaves was determined by real-time PCR analysis and normalized to the plant RPS18B mRNA level. The fold change was calculated relative to control which was mechanical wounding + H2O. The graph shows x-fold induction of the mRNA levels by the S. littoralis OS relative to the levels in the H2O treated control leaves. Mean (± SE, n = 5). Different letters indicate significant differences between different time points in a single gene expression (ANOVA; p < 0.05). Comparisons between expressions of different genes are not performed.
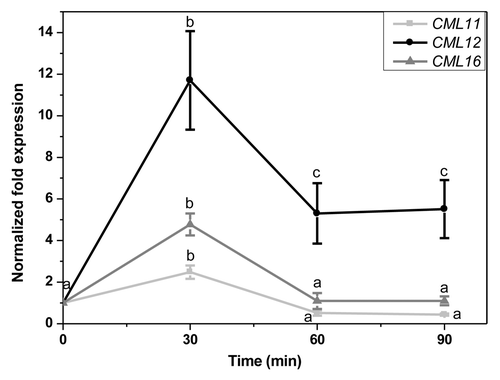
Figure 2.CML9, CML17, and CML23 transcript levels in leaves of A. thaliana 30, 60 and 90 min after treatment with Spodoptera littoralis oral secretion (OS). Leaves were elicited by pattern wheel wounding and subsequently treating the wound with 20 µL water or 1:1 diluted OS per leaf. Transcript abundance in leaves was determined by real-time PCR analysis and normalized to the plant RPS18B mRNA level. The fold change was calculated relative to control which was mechanical wounding + H2O. The graph shows x-fold induction of the mRNA levels by the S. littoralis OS relative to the levels in the H2O treated control leaves. Mean (± SE, n = 3). Different letters indicate significant differences between different time points in a single gene expression (ANOVA; p < 0.05). Comparisons between expressions of different genes are not performed.
To investigate and finally understand the specific roles of the identified CMLs, in plant herbivore interactions, further functional experiments involving knockout and overexpression of target genes are necessary. However, the fact that at least seven CMLs are regulated on the expression level by application of OS and herbivory suggests a central function for these calcium sensors in plant defense signaling processes
Materials and Methods
Plant growth and treatment
Arabidopsis thaliana seeds (ecotype Columbia) were used for all experiments and grown as described.Citation12 Experiments with insect oral secretions (OS) were performed according to.Citation12 Briefly, wounding was done with a pattern wheel (6 vertical motions) on either side of the leaf. OS was collected from 4th instar Spodoptera littoralis larvae reared on artificial diet and fed on A. thaliana leaves for 24 h prior to OS collection. The harvested OS was centrifuged for 2 min at 13,000 rpm and subsequently diluted 1:1 with water. A total of 20 µl of fresh diluted OS was spread across all the holes on a single leaf. In control plants, water was added. The samples were harvested and stored in liquid nitrogen. Experiments were repeated three times independently.
Expression analysis by Real Time PCR
Leaf material was ground to a fine powder in liquid N2, and total RNA was isolated using the TRIzol Reagent (Invitrogen) according to the manufacturers´ protocol. An additional DNase (Turbo DNase, Ambion) treatment was included to eliminate any contaminating DNA. RNA quantity was determined photospectrometrically. Total RNA (1 µg) was converted into single-stranded cDNA using a mix of oligo-dT20 primers using the Omniscript cDNA synthesis kit (Qiagen). Gene-specific primers were designed using the NCBI primer design tool (http://www.ncbi.nlm.nih.gov/tools/primer-blast). For real time PCR, primers producing 124 to 190 bp amplicons were used. Q-RT-PCR was done in optical 96-well plates on a MX3000P Real-Time PCR Detection System (Stratagene) using the Brilliant II QPCR SYBR green Mix (Agilent) to monitor double-stranded DNA synthesis in combination with ROX as a passive reference dye included in the PCR master mix. A dissociation curve analysis was performed for all primer pairs, and all experimental samples yielded a single sharp peak at the amplicon’s melting temperature. The mRNA levels for each cDNA probe were normalized with respect to the RPS18B mRNA level.Citation12 Fold induction values of target genes were calculated with the ΔΔCP equationCitation20 and normalized to the mRNA level of target genes in control leaf, which were defined as 1.0. All of the assays were run in triplicate (biological replication) to control for overall variability. Primer pairs (forward, reverse) used are listed below:
CML9 (At3g51920):
5′- TTGGCAACGGTGGCATCACT -3′
5′- CCATCGCCATCAAGGTCGGCT -3′
CML11 (At3g22930):
5′- TCCGCTCATTGGATCAGAACCCT -3′
5′- TCTGCATCAGTTTCCTGGAGTTGGT -3′
CML12- (At2g41100):
5′- TGGCGGATAAGCTCACTGACGA -3′
5′- TCCGCTTCGTTCATCAAGTCCTG -3′
CML16 (At3g25600):
5′- GACGAGCTGGTCGTGGCGAT -3′
5′- TGACCCAGCAAGTTCCGCCG -3′
CML17 (At1g32250):
5′- CGCCGGCGAAGAGGACAACT -3′
5′- ATTCCGCCACCGTCAAGGCG -3′
CML23 (At1g66400):
5′- CGCTTCACAAGAAGAAACCAAAGCA -3′
5′- AGCCGAGATCCTTCCATTACGATCC -3′
RPS18B (At1g34030):
5′- GTCTCCAATGCCCTTGACAT -3′
5′- TCTTTCCTCTGCGACCAGTT -3′
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank A. Berg for rearing caterpillars, the greenhouse team for growing plants, W. Boland for continuous support, and the Max Planck Society for funding.
References
- Kudla J, Batistic O, Hashimoto K. Calcium signals: the lead currency of plant information processing. Plant Cell 2010; 22:541 - 63; http://dx.doi.org/10.1105/tpc.109.072686; PMID: 20354197
- Dodd AN, Kudla J, Sanders D. The language of calcium signaling. Annu Rev Plant Biol 2010; 61:593 - 620; http://dx.doi.org/10.1146/annurev-arplant-070109-104628; PMID: 20192754
- Sanders D, Pelloux J, Brownlee C, Harper JF. Calcium at the crossroads of signaling. Plant Cell 2002; 14:Suppl S401 - 17; PMID: 12045291
- Perochon A, Aldon D, Galaud J-P, Ranty B. Calmodulin and calmodulin-like proteins in plant calcium signaling. Biochimie 2011; 93:2048 - 53; http://dx.doi.org/10.1016/j.biochi.2011.07.012; PMID: 21798306
- McCormack E, Tsai YC, Braam J. Handling calcium signaling: Arabidopsis CaMs and CMLs. Trends Plant Sci 2005; 10:383 - 9; http://dx.doi.org/10.1016/j.tplants.2005.07.001; PMID: 16023399
- Delk NA, Johnson KA, Chowdhury NI, Braam J. CML24, regulated in expression by diverse stimuli, encodes a potential Ca2+ sensor that functions in responses to abscisic acid, daylength, and ion stress. Plant Physiol 2005; 139:240 - 53; http://dx.doi.org/10.1104/pp.105.062612; PMID: 16113225
- Hubbard K, Hotta C, Gardner M, Braam J, Webb A. The Arabidopsis thaliana calmodulin-like protein CML24 is a regulator of rhythmic Ca2+ signalling and flowering time. Comp Biochem Physiol A Mol Integr Physiol 2008; 150:S153 - 153; http://dx.doi.org/10.1016/j.cbpa.2008.04.395
- Vanderbeld B, Snedden WA. Developmental and stimulus-induced expression patterns of Arabidopsis calmodulin-like genes CML37, CML38 and CML39.. Plant Mol Biol 2007; 64:683 - 97; http://dx.doi.org/10.1007/s11103-007-9189-0; PMID: 17579812
- Park HC, Park CY, Koo SC, Cheong MS, Kim KE, Kim MC, et al. AtCML8, a calmodulin-like protein, differentially activating CaM-dependent enzymes in Arabidopsis thaliana. Plant Cell Rep 2010; 29:1297 - 304; http://dx.doi.org/10.1007/s00299-010-0916-7; PMID: 20820784
- Magnan F, Ranty B, Charpenteau M, Sotta B, Galaud J-P, Aldon D. Mutations in AtCML9, a calmodulin-like protein from Arabidopsis thaliana, alter plant responses to abiotic stress and abscisic acid. Plant J 2008; 56:575 - 89; http://dx.doi.org/10.1111/j.1365-313X.2008.03622.x; PMID: 18643966
- Leba L-J, Cheval C, Ortiz-Martín I, Ranty B, Beuzón CR, Galaud J-P, et al. CML9, an Arabidopsis calmodulin-like protein, contributes to plant innate immunity through a flagellin-dependent signalling pathway. Plant J 2012; •••; http://dx.doi.org/10.1111/j.1365-313X.2012.05045.x; PMID: 22563930
- Vadassery J, Reichelt M, Hause B, Gershenzon J, Boland W, Mithöfer A. CML42-mediated calcium signaling co-ordinates responses to Spodoptera herbivory and abiotic stresses in Arabidopsis.. Plant Physiol 2012; 159:1159 - 75; http://dx.doi.org/10.1104/pp.112.198150; PMID: 22570470
- Dobney S, Chiasson D, Lam P, Smith SP, Snedden WA. The calmodulin-related calcium sensor CML42 plays a role in trichome branching. J Biol Chem 2009; 284:31647 - 57; http://dx.doi.org/10.1074/jbc.M109.056770; PMID: 19720824
- Braam J, Davis RW. Rain-, wind-, and touch-induced expression of calmodulin and calmodulin-related genes in Arabidopsis. Cell 1990; 60:357 - 64; http://dx.doi.org/10.1016/0092-8674(90)90587-5; PMID: 2302732
- Sistrunk ML, Antosiewicz DM, Purugganan MM, Braam J. Arabidopsis TCH3 encodes a novel Ca2+ binding protein and shows environmentally induced and tissue-specific regulation. Plant Cell 1994; 6:1553 - 65; PMID: 7827491
- Braam J. Regulated expression of the calmodulin-related TCH genes in cultured Arabidopsis cells: induction by calcium and heat shock. Proc Natl Acad Sci U S A 1992; 89:3213 - 6; http://dx.doi.org/10.1073/pnas.89.8.3213; PMID: 1373491
- Antosiewicz DM, Polisensky DH, Braam J. Cellular localization of the Ca2+ binding TCH3 protein of Arabidopsis. Plant J 1995; 8:623 - 36; http://dx.doi.org/10.1046/j.1365-313X.1995.08050623.x; PMID: 8528275
- Polisensky DH, Braam J. Cold-shock regulation of the Arabidopsis TCH genes and the effects of modulating intracellular calcium levels. Plant Physiol 1996; 111:1271 - 9; http://dx.doi.org/10.1104/pp.111.4.1271; PMID: 8756505
- Tsai YC, Delk NA, Chowdhury NI, Braam J. Arabidopsis potential calcium sensors regulate nitric oxide levels and the transition to flowering. Plant Signal Behav 2007; 2:446 - 54; http://dx.doi.org/10.4161/psb.2.6.4695; PMID: 19517005
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 2001; 29:e45; http://dx.doi.org/10.1093/nar/29.9.e45; PMID: 11328886