Abstract
In response to plant proximity or canopy shade, plants can react by altering elongation growth and development. Several members of the class II homeodomain-leucine zipper (HD-ZIPII) transcription factor family have been shown to play an instrumental role in the responses to shade. HD-ZIP members of the class III (HD-ZIPIII), by contrast, are involved in basic patterning processes. We recently showed that REVOLUTA (REV), a member of the HD-ZIPIII family, directly and positively regulates the expression of several genes involved in shade-induced growth, such as those encoding HD-ZIPII factors HAT2, HAT3, ATHB2/HAT4 and ATHB4, and of the components of the auxin biosynthesis pathway YUCCA5 and TAA1. Furthermore, we could demonstrate a novel role for HD-ZIPIII in shade-induced promotion of growth. Here we show that besides responding to shade, ATHB4 and HAT3 have a critical role in establishing the dorso-ventral axis in cotyledons and developing leaves. Loss-of-function mutations in these two HD-ZIPII genes (athb4 hat3) results in severely abaxialized, entirely radialized leaves. Conversely, overexpression of HAT3 results in adaxialized leaf development. Taken together, our findings unravel a so far unappreciated role for an HD-ZIPII/HD-ZIPIII module required for dorso-ventral patterning of leaves. The finding that HD-ZIPII/HD-ZIPIII also function in shade avoidance suggests that this module is at the nexus of patterning and growth promotion.
Introduction
Plants are sessile organisms and to maximize reproductive success, they have to adjust their growth behavior to their environment. Light is one of the most important environmental cues as it provides both energy and information. Plants have evolved refined mechanisms to detect both light quality and quantity and to measure the duration of the light period. Important growth responses and developmental decisions, such as plant architecture and the transition to flowering, are influenced by a combination of cues such as light quality and day length. In nature, plants live in communities with other plant species that might compete for resources. To avoid living under a canopy, they can detect plant proximity and canopy shade as changes in the red (R) to far-red (FR) ratio (R:FR ratio) of light and translate these changes into growth responses, collectively known as the shade avoidance syndrome (SAS) that include enhanced hypocotyl elongation, reduced leaf expansion, decreased branching and accelerated flowering.Citation1-Citation3 As plant leaves reflect FR-light, neighboring plants can sense subtle decreases in R:FR ratio (‘neighbor proximity detection’) and react by inducing hypocotyl growth.Citation4 In case of true plant shade, canopy plant leaves selectively absorb light from the photosynthetic active radiation, which includes R light. Therefore, both the R:FR and the overall quantity of the photosynthetic active radiation (400–700 nm) is decreased (canopy shade conditions), which is also translated into growth-induction of the hypocotyl.Citation1-Citation3 Changes in R:FR are perceived by the plant phytochrome system, which rapidly influences hormonal responses and a downstream transcriptional network to alter the mentioned aspects of plant development and architecture.Citation5-Citation7
The transcriptome of Arabidopsis changes significantly in response to shadeCitation8,Citation9 and numerous shade-induced genes are known.Citation10,Citation11 Several of these rapidly shade induced genes belong to the class II homeodomain leucine-zipper (HD-ZIPII) family of transcription factors, that are mostly known to be involved in the regulation of adaptive responses to the environment.Citation12,Citation13 The recent finding that HAT1/JAIBA, a HD-ZIPII protein, is also involved in the regulation of meristem activityCitation14 hints toward additional functions of HD-ZIPIIs such as regulation of plant development per se. We could recently show that the expression of several HD-ZIPII genes is directly controlled by the HD-ZIPIII transcription factor REVOLUTA (REV).Citation15 HD-ZIPIII factors have known roles in controlling embryo, shoot and root patterningCitation16-Citation18 and our previous finding that they are involved in an adaptive process such as the SAS, suggested that they function at the nexus of adjusting growth to the environment. Previously it was shown that double mutant plants in two HD-ZIPII genes (ATHB4 and HAT3) display strong alterations in their development.Citation3,Citation10 Using a genetic approach, we have investigated whether HD-ZIPII transcription factors also have a role in the regulation of leaf development. Our data demonstrates that the combined loss of ATHB4 and HAT3 function results in radialized leaves with abaxial characteristics, reminiscent of hd-zipIII mutant plants. Conversely, the analysis of gain-of-function overexpression plants reveals that HAT3 promotes adaxial leaf development, strongly resembling HD-ZIPIII overexpression. Taken together our results support a role for these two HD-ZIPII factors downstream of HD-ZIPIII in the regulation of leaf development. Since long-term exposure to shade results in leaves with longer petioles but reduced leaf blades,Citation3,Citation19 our results suggest that HD-ZIPs might be part of the mechanisms regulating this process.
Results and Discussion
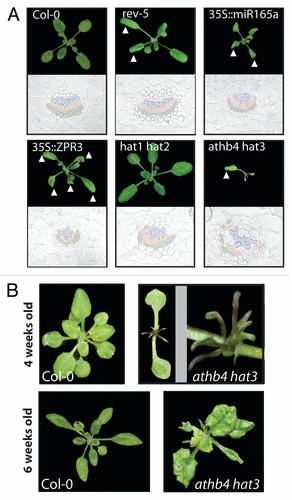
Our analysis of REV target genes revealed several HD-ZIPII transcription factors that are directly and positively regulated by REV.Citation15 Some HD-ZIPII transcription factors are known to be involved in shade signalingCitation10 and our recent analysis showed that HD-ZIPIIIs are also involved in shade growth ().Citation15 Using available double mutant plants in HD-ZIPII genes (hat1 hat2 and athb4 hat3) and various hd-zipIII mutant plants (rev-5, 35S::miR165a and 35S::ZPR3), we performed comparative leaf growth studies. When grown side-by-side, athb4 hat3 double mutant plants were severely impaired in development and retarded in growth before reaching the reproductive phase (). The hat1 hat2 double mutant did not display a mutant phenotype in regard to altered leaf polarity (). Together, these studies revealed that, like HD-ZIPIIIs, also some HD-ZIPIIs play a prominent role in regulating polar leaf development in Arabidopsis. We next examined the vascular strands of petioles of different hd-zipII/hd-zipIII mutant plants to detect more subtle polarity-associated defects. Vascular strands of wild type plants, as well as hat1 hat2 double mutant, showed a typical sandwich-structure with xylem on top (colored in blue), cambium cells in the middle (colored in red) and phloem on the bottom (green). In plants with reduced HD-ZIPIII activity (35S::miR165a, rev-5, 35S::ZPR3), the vasculature showed different degrees of abaxialized and radialized characteristics, with phloem surrounding the xylem. Histological analyses of vascular strands of leaves of athb4 hat3 double mutant plants showed strong abaxialization, manifested by radialization of transport elements and also a severe disruption of the overall organization (). Thus, the athb4 hat3 mutant phenotype somewhat resembles hd-zipIII mutant plants. Interestingly, in our growth conditions, the mutations caused strong leaf patterning defects in the early post-embryonic growth phase and both cotyledons and early leaves showed strong developmental defects (). Later in development, the mutant athb4 hat3 phenotype was alleviated and leaf development resumed to a more normal state (, lower panel), an effect that was not observed in hd-zipIII mutant plants. These findings illustrate that these two HD-ZIPII transcription factors play an important role in leaf patterning, very likely downstream of HD-ZIPIII action. Because they affect more strongly the early post-embryonic growth phase, their action might be less required for the development leaves formed by older plants.
Figure 1. Model showing the role of REV targets in shade avoidance and leaf development. Using a ChIP-Seq approach we have previously shown that REV acts upstream of both HD-ZIPII and auxin biosynthesis. Thereby REV influences shade-induced growth responses. Here we show that HD-ZIPII transcription factors also have a prominent role in regulating leaf development. It is unknown whether and how HD-ZIPIIIs are activated by shade and whether TAA1/YUC5 play a role in leaf development.
Figure 2. Mutations in both HD-ZIPII and HD-ZIPIII genes cause polarity defects in leaves and vascular organization. (A) Comparative growth analysis of different mutant plants. The triangle highlights abaxialized leaves of rev-5, 35S::miR165a, 35S::ZPR3 and athb4 hat3 plants. In wild type Col-0 and hat1 hat2 plants no growth abnormalities were observed. Below the photographs of wild type and the different mutant plants, sections through petioles are shown. The vasculature of wild type Col-0 plants shows the typical sandwich structure tissue containing phloem cells (green), cambium cells (red) and tissue containing xylem elements (blue) on top. Both 35S::miR165a and 35S::ZPR3 transgenic plants show abaxialized vascular strands with phloem nearly surrounding the xylem. In athb4 hat3 mutant plants, the vascular organization is severely disturbed but is also showing abaxialized characteristics. (B) In the juvenile phase of post-embryonic growth, athb4 hat3 double mutants produce radial leaves in comparison to Col-0 wild type plants. Later in development (around 6 weeks after germination; lower panel), leaves with weaker abaxialized characteristics, such as downward bending leaf blade, are being produced.
Using scanning-electron microscopy we further characterized the early growth defects of athb4 hat3 mutant plants and could observe that both cotyledons and leaves were radialized to different degrees and lacked adaxial characteristics () compared with wild type plants (). Wild type and athb4 hat3 mutant seedlings were also compared using confocal microscopy and 3D-reconstruction (, Vids. S1 and S2), displaying the alterations in leaf development at higher resolution. Optical sections through developing cotyledons revealed normal polarity of wild-type cotyledons with vascular strands vs. strongly radialized and abaxialized cotyledons with disorganized vascular strands in athb4 hat3 mutant plants (). The results further corroborated that ATHB4 and HAT3 transcription factors are involved in patterning the adaxial domain in the early leaf primordium.
Figure 3. Mutations in HD-ZIPII genes cause abnormal growth and polarity defects in cotyledons and leaves. (A-D) Scanning Electron Microcopy images showing wild-type and athb4 hat3 mutant seedlings on days 3 and 5. Phenotypes of wild type (A, C, E, G) and athb4 hat3 (B, D, F, H) seedlings on day 3 (cotyledons: A, B), day 5 (primary leaves: B, D), day 7 (cotyledons: E, F) and day 10 (primary leaves: G, H). (I) Confocal microscopy images showing a transversal section of a wild-type (left) and athb4 hat3 (right) cotyledon to visualize the different types of cells. (J) 3D-reconstruction using the Osirix software from image sections of wild-type (left) and athb4 hat3 (right) seedlings. Images were taken from 1-d-old seedlings. Mutant seedlings lack mostly palisade cells, suggesting they are missing adaxial identity. Bar size corresponds to 0.5 (A, B), 0.2 (C, D), 1 (E, F) and 0.3 (G, H) mm. (K) Real-time quantitative PCR (qPCR) experiments showing expression changes of PHB and PHV in Col-0 and athb4 hat3 mutant seedlings. Transcript abundance is measured relative to Col-0 values. Values are means ± SE of three (Col-0) or five (athb4 hat3) independent biological qPCR replicates normalized to UBQ10. Asterisk indicate significant differences (p < 0.01) relative to the Col-0 plants growing under the same conditions.
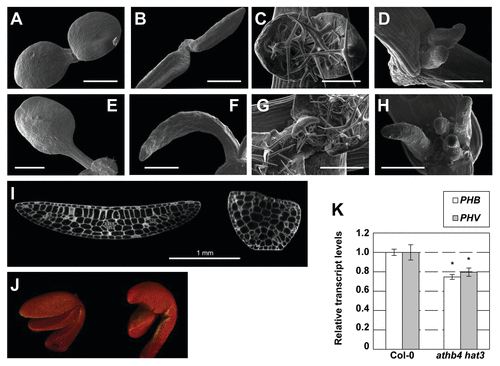
To find out whether mis-regulation of HD-ZIPIII genes is a consequence of the athb4 hat3 mutant phenotype, we analyzed the expression PHB and PHV, two adaxial marker genes of the HD-ZIPIII family. Plants carrying dominant mutations in either PHB or PHV display dramatic adaxialized phenotypesCitation17 and thus behave opposite to the developmental defects observed in athb4 hat3 mutant plants. Our expression analysis shows that both PHB and PHV expression is significantly lower in athb4 hat3 mutant plants compared with Col-0 wild type plants (). These findings suggest that besides acting downstream of REV, ATHB4 and HAT3 might have an additional function upstream of REV. The observation that PHB and PHV expression are reduced in athb4 hat3 mutant plants might suggest that ATHB4 and HAT3 act positively on HD-ZIPIII expression. Based on the auto-activation capacity in a yeast two-hybrid assay, it has been suggested that HAT1/JAIBA may act as a transcriptional activator.Citation14 However, all HD-ZIPII proteins contain an N-terminal EAR motif, required for transcriptional repression.Citation20 It is furthermore known that when overexpressed in plants, several HD-ZIPIIs act as transcriptional repressors over the expression of some genes,Citation10,Citation21-Citation23 for which reason it is unlikely that they act by directly and positively regulating HD-ZIPIII expression. Therefore it seems plausible that the reduced expression of PHB and PHV is an indirect effect, i.e., a mere consequence of reduced adaxial tissue. We therefore conclude that the combined loss of ATHB4 and HAT3 causes strongly abaxialized leaf development, which is reflected by reduced expression of the adaxial identity markers PHB and PHV.
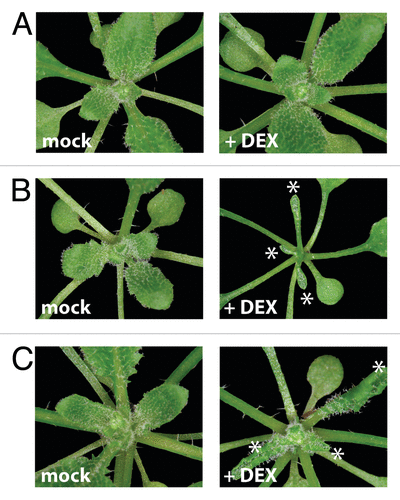
We next examined whether the ectopic expression of HAT3, a REV target gene, can elicit phenotypes associated with either loss- or gain-of-REV function. In order to avoid strong pleiotropic overexpression-phenotypes, we decided to employ the glucocorticoid-receptor (GR) inducible system. Using Gateway recombination (Invitrogen), transgenic 35S::FLAG-GR-HAT3 plants were constructed. Four-week old transgenic T2 plants (n = 20) were grown in short day conditions and treated once a day with DEX by spraying for one week. DEX-induced transgenic 35S::FLAG-GR-HAT3 plants showed strong upward-curling of leaf blades, largely resembling DEX-induced 35S::FLAG-GR-REVd transgenic plants (), a phenotype caused by over-proliferation of adaxial-derived tissue in leaves.Citation24 These data further support our previous conclusion that HD-ZIPIIIs and these two HD-ZIPIIs (ATHB4 and HAT3) have common functions in both shade-induced growth promotion and leaf patterning.
Figure 4. Inducible overexpression lines of either REVOLUTA or HAT3 cause similar developmental alterations in leaf formation. Four-week old soil-grown seedlings treated for one week with either Dexamethasone or a mock solution by spraying. Strongly adaxialized leaves (see asterisks) were observed in DEX-treated 35S::FLAG-GR-HAT3 (B) and 35S::FLAG-GR-REVd(C) plants in comparison to Col-0 wild type plants (A).
Besides promotion of hypocotyl growth, long-term exposure to shade significantly alters the leaf developmental program.Citation19 A reduced leaf blade expansion has been commonly observed in plants that constitutively overexpressed some HD-ZIPII, such as ATHB2/HAT4,Citation25 ATHB4,Citation10 HAT2Citation21 or HAT3 (), suggesting that high levels of HD-ZIPII transcription factors might account for this SAS-related phenotype. Our results suggested that HD-ZIPIIIs might also participate in this SAS response. It remains unclear how HD-ZIPIIs execute their influence on dorso-ventral axis formation. It seems conceivable that both HD-ZIPII and HD-ZIPIII act in a common protein complex, which, based on environmental influence, switches between transcriptional activation and repression. Alternatively, HD-ZIPIIIs can activate the expression of HD-ZIPIIs-encoding genes in concert with environmental conditions, which in turn would repress expression of the actual executors involved in dorso-ventral leaf patterning.
Materials and Methods
Fixation, clearing and staining procedure for three-dimensional imaging
For the experiments shown in , seeds were germinated and grown in a growth chamber at 22°C under continuous white light (W, 80 μmol·m−2 s−1 of photosynthetic active radiation; R:FR ratio of 3.2–4.5). On the day of harvest, plant material was processed as described previously.Citation26-Citation28 Briefly, seedlings of different ages were immersed in fixative solution (50% methanol and 10% acetic acid) at 4°C for at least 24 h (up to 1 mo). Plant material was then transferred to 80% (v/v) ethanol and incubated at 80°C for 5 min, briefly rinsed in ethanol dilutions (70%, 50%, 30% and 10% (v/v) ethanol) and finally rinsed twice with water. Then samples were incubated in 1% (v/v) periodic acid at room temperature with gentle agitation (about 100 rpm) for 40 min, rinsed again with water and incubated in Schiff reagent with freshly prepared 100 mg/mL propidium iodide in 100 mM sodium metabisulphite and 0.15 N HCl for 1–2 h (until plants were visibly stained). Then samples were rinsed with water and transferred onto microscope slides and covered with chloral hydrate solution (4 g chloral hydrate, 1 mL glycerol, and 2 mL water). Slides were kept overnight at room temperature in a closed environment to prevent drying out. The following day, the excess of chloral hydrate was removed, several drops of Hoyer’s solution (30 g gum arabic, 200 g chloral hydrate, 20 g glycerol and 50 mL water) were added and a coverslip was placed on top. Slides were left undisturbed for a minimum of 3 d to allow the mounting solution to set.
Confocal microscopy and data processing
A Leica TCS-SP2-AOBS spectral confocal laser-scanning microscope (Leica Microsystems) was used. The excitation wavelength for PS-PI–stained samples was 488 nm, and emission was collected at 520 to 720 nm. Data were processed for some two-dimensional orthogonal sections, 3D rendering and movie exports using the open source software OsirixCitation29(http://osirix.softonic.com/mac/) on a quadxeon 2.66-Ghz, 2-GB RAM Apple Mac pro workstation. RGB stacks of confocal images were imported as DICOM files into Osirix prior to surface rendering.
Scanning electron microscopy
For scanning electron microscopy, seeds were sown on growth medium containing 1% (w/v) sucrose. After stratification (3 d), plates were transferred to continuous white light. On days 0, 3, 5, 7 and 10, plant material was transferred into the microscope without any further treatment. Plant material was imaged with a MEB Hirox SH-1500 (Hirox Europe-Jyfel) microscope at -30°C.
RNA expression analysis by quantitative PCR
For reverse transcriptase quantitative PCR (qPCR) analyses of gene expression, seeds were sown on filter paper on top of GM- medium. Seedlings were grown under continuous W for 7 d. qPCR analyses were performed as indicated elsewhere.10UBQ10 gene was used for normalization. We assayed 3–5 biological replicas for each sample. Primer sequences for qPCR were MSO40 (5′- GCT AAC AAC CCA GCA GGA CTC CT -3′) and MSO41 (5′- TAA GCT CGA TCG TCC CAC CGT T -3′) for PHB (At2g34710) and MSO42 (5′- GCT AAT CTT CTC TCG ATT GCG GAG GA -3′) and MSO43 (5′- GCT CGA TAG TAC CAC CAT TTC CAG TG -3′) for PHV (At1g30490). Primers for UBQ10 transcript level analyses were described before.Citation10
| Abbreviations: | ||
| HD-ZIP | = | homeodomain leucine-zipper |
| R:FR ratio | = | red to far-red ratio |
Additional material
Download Zip (31.9 KB)Acknowledgments
The excellent technical support from GesineSeibold and Ingrid Blumberg is gratefully acknowledged. Research in the J.F.M.-G. lab is supported by grants from the Generalitat de Catalunya (Xarba, 2009-SGR697) and MICINN - FEDER funds (CSD2007–00036, BIO2008–00169, BIO2011–23489). Research in the S.W. laboratory is made possible by funding from the Deutsche Forschungsgemeinschaft (DFG, WE4281/6–1), the European Union (FP7 No. 256502), the Robert Bosch Foundation and the Ministry for Agriculture (BundesanstaltfürLandwirtschaft und Ernährung; CHN13–22/12–13).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Ballaré CL. Keeping up with the neighbours: phytochrome sensing and other signalling mechanisms. Trends Plant Sci 1999; 4:97 - 102; 10.1016/S1360-1385(99)01383-7; PMID: 10322540
- Franklin KA. Shade avoidance. New Phytol 2008; 179:930 - 44; 10.1111/j.1469-8137.2008.02507.x; PMID: 18537892
- Martinez-Garcia JF, Galstyan A, Salla-Martret M, Cifuentes-Esquivel N, Gallemi M, Bou-Torrent J. Regulatory Components of Shade Avoidance Syndrome. Advances in Botanical Research 2010; 53:65 - 116; 10.1016/S0065-2296(10)53003-9
- Ballaré CL, Scopel AL, Sánchez RA. Far-red radiation reflected from adjacent leaves: an early signal of competition in plant canopies. Science 1990; 247:329 - 32; 10.1126/science.247.4940.329; PMID: 17735851
- Li L, Ljung K, Breton G, Schmitz RJ, Pruneda-Paz J, Cowing-Zitron C, et al. Linking photoreceptor excitation to changes in plant architecture. Genes Dev 2012; 26:785 - 90; 10.1101/gad.187849.112; PMID: 22508725
- Roig-Villanova I, Bou-Torrent J, Galstyan A, Carretero-Paulet L, Portolés S, Rodríguez-Concepción M, et al. Interaction of shade avoidance and auxin responses: a role for two novel atypical bHLH proteins. EMBO J 2007; 26:4756 - 67; 10.1038/sj.emboj.7601890; PMID: 17948056
- Hornitschek P, Kohnen MV, Lorrain S, Rougemont J, Ljung K, López-Vidriero I, et al. Phytochrome interacting factors 4 and 5 control seedling growth in changing light conditions by directly controlling auxin signaling. Plant J 2012; In Press 10.1111/j.1365-313X.2012.05033.x; PMID: 22536829
- Sessa G, Carabelli M, Sassi M, Ciolfi A, Possenti M, Mittempergher F, et al. A dynamic balance between gene activation and repression regulates the shade avoidance response in Arabidopsis. Genes Dev 2005; 19:2811 - 5; 10.1101/gad.364005; PMID: 16322556
- Tao Y, Ferrer JL, Ljung K, Pojer F, Hong FX, Long JA, et al. Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 2008; 133:164 - 76; 10.1016/j.cell.2008.01.049; PMID: 18394996
- Sorin C, Salla-Martret M, Bou-Torrent J, Roig-Villanova I, Martínez-García JF. ATHB4, a regulator of shade avoidance, modulates hormone response in Arabidopsis seedlings. Plant J 2009; 59:266 - 77; 10.1111/j.1365-313X.2009.03866.x; PMID: 19392702
- Roig-Villanova I, Bou J, Sorin C, Devlin PF, Martínez-García JF. Identification of primary target genes of phytochrome signaling. Early transcriptional control during shade avoidance responses in Arabidopsis. Plant Physiol 2006; 141:85 - 96; 10.1104/pp.105.076331; PMID: 16565297
- Ariel FD, Manavella PA, Dezar CA, Chan RL. The true story of the HD-Zip family. Trends Plant Sci 2007; 12:419 - 26; 10.1016/j.tplants.2007.08.003; PMID: 17698401
- Ciarbelli AR, Ciolfi A, Salvucci S, Ruzza V, Possenti M, Carabelli M, et al. The Arabidopsis homeodomain-leucine zipper II gene family: diversity and redundancy. Plant Mol Biol 2008; 68:465 - 78; 10.1007/s11103-008-9383-8; PMID: 18758690
- Zúñiga-Mayo VM, Marsch-Martínez N, de Folter S. JAIBA, a class-II HD-ZIP transcription factor involved in the regulation of meristematic activity, and important for correct gynoecium and fruit development in Arabidopsis. Plant J 2012; 71:314 - 26; 10.1111/j.1365-313X.2012.04990.x; PMID: 22409594
- Brandt R, Salla-Martret M, Bou-Torrent J, Musielak T, Stahl M, Lanz C, et al. Genome-wide binding-site analysis of REVOLUTA reveals a link between leaf patterning and light-mediated growth responses. Plant J 2012; In press 10.1111/j.1365-313X.2012.05049.x; PMID: 22578006
- Carlsbecker A, Lee JY, Roberts CJ, Dettmer J, Lehesranta S, Zhou J, et al. Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate. Nature 2010; 465:316 - 21; 10.1038/nature08977; PMID: 20410882
- McConnell JR, Emery J, Eshed Y, Bao N, Bowman J, Barton MK. Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 2001; 411:709 - 13; 10.1038/35079635; PMID: 11395776
- Smith ZR, Long JA. Control of Arabidopsis apical-basal embryo polarity by antagonistic transcription factors. Nature 2010; 464:423 - 6; 10.1038/nature08843; PMID: 20190735
- Morelli G, Ruberti I. Light and shade in the photocontrol of Arabidopsis growth. Trends Plant Sci 2002; 7:399 - 404; 10.1016/S1360-1385(02)02314-2; PMID: 12234731
- Kagale S, Links MG, Rozwadowski K. Genome-wide analysis of ethylene-responsive element binding factor-associated amphiphilic repression motif-containing transcriptional regulators in Arabidopsis. Plant Physiol 2010; 152:1109 - 34; 10.1104/pp.109.151704; PMID: 20097792
- Sawa S, Ohgishi M, Goda H, Higuchi K, Shimada Y, Yoshida S, et al. The HAT2 gene, a member of the HD-Zip gene family, isolated as an auxin inducible gene by DNA microarray screening, affects auxin response in Arabidopsis. Plant J 2002; 32:1011 - 22; 10.1046/j.1365-313X.2002.01488.x; PMID: 12492842
- Ohgishi M, Oka A, Morelli G, Ruberti I, Aoyama T. Negative autoregulation of the Arabidopsis homeobox gene ATHB-2. Plant J 2001; 25:389 - 98; 10.1046/j.1365-313x.2001.00966.x; PMID: 11260495
- Steindler C, Matteucci A, Sessa G, Weimar T, Ohgishi M, Aoyama T, et al. Shade avoidance responses are mediated by the ATHB-2 HD-zip protein, a negative regulator of gene expression. Development 1999; 126:4235 - 45; PMID: 10477292
- Wenkel S, Emery J, Hou B-H, Evans MMS, Barton MK. A feedback regulatory module formed by LITTLE ZIPPER and HD-ZIPIII genes. Plant Cell 2007; 19:3379 - 90; 10.1105/tpc.107.055772; PMID: 18055602
- Schena M, Lloyd AM, Davis RW. The HAT4 gene of Arabidopsis encodes a developmental regulator. Genes Dev 1993; 7:367 - 79; 10.1101/gad.7.3.367; PMID: 8449400
- Wuyts N, Palauqui J-C, Conejero G, Verdeil J-L, Granier C, Massonnet C. High-contrast three-dimensional imaging of the Arabidopsis leaf enables the analysis of cell dimensions in the epidermis and mesophyll. Plant Methods 2010; 6:17; 10.1186/1746-4811-6-17; PMID: 20598116
- Truernit E, Bauby H, Dubreucq B, Grandjean O, Runions J, Barthélémy J, et al. High-resolution whole-mount imaging of three-dimensional tissue organization and gene expression enables the study of Phloem development and structure in Arabidopsis. Plant Cell 2008; 20:1494 - 503; 10.1105/tpc.107.056069; PMID: 18523061
- Truernit E, Palauqui J-C. Looking deeper: whole-mount confocal imaging of plant tissue for the accurate study of inner tissue layers. Plant Signal Behav 2009; 4:151 - 2; 10.4161/psb.4.2.7683; PMID: 19649197
- Rosset A, Spadola L, Ratib O. OsiriX: an open-source software for navigating in multidimensional DICOM images. J Digit Imaging 2004; 17:205 - 16; 10.1007/s10278-004-1014-6; PMID: 15534753