Abstract
2-Hydroxy fatty acids mainly contained in sphingolipids are synthesized by a sphingolipid fatty acid 2-hydroxylase (FAH). Recently, two FAH homologs in Arabidopsis thaliana (AtFAH1 and AtFAH2), without any cytochrome b5(Cb5)-like domains, which are essential for the function of Saccharomyces cerevisiae and mammalian FAH, were identified and both AtFAHs were shown to be activated by the interaction with Cb5. In this study, we compared FAHs of various plants, animals and fungi. Interestingly, only plants had two FAH homologs and none of plant FAHs had any Cb5-like domains. In addition, we showed from the interaction and expression analyses that AtFAHs interacted with multiple Cb5s probably in various tissues. Thus, plant FAHs may have evolved unlike animal and fungus FAHs.
Text
In eukaryotes, 2-hydroxy fatty acids (2-HFAs) are special fatty acids whose C-2 position is hydroxylated and that are contained in only particular lipids such as sphingolipids.Citation1-Citation3In vitro biophysical works have demonstrated that the 2-hydroxy group in sphingolipid fatty acids plays a crucial role in the lipid organization within membranes for its hydrogen bonding capability.Citation4-Citation6
The 2-hydroxylation of sphingolipid N-acyl chains is catalyzed by the enzyme sphingolipid fatty acid 2-hydroxylase (FAH).Citation3 Since FAH was first identified in Saccharomyces cerevisiae (S. cerevisiae), FAH homologs have been found in mammals (FA2H) such as human, mouse and rat.Citation3,Citation7-Citation9 S. cerevisiae and mammalian FAHs are endoplasmic reticulum (ER)-localized membrane proteins with an N-terminal cytochrome b5 (Cb5)-like domain essential for its enzymatic activity. FAH also contains several consensus histidine motifs as a catalytic site. These histidine residues coordinate the nonheme di-iron cluster (Fe-O-Fe) at the active site of the enzyme and receive electrons from its Cb5-like domain. Recent vigorous studies have revealed that the loss of mammalian FA2H is related to serious diseases such as spastic paraplegia, leukodystrophy, brain ion deposition and alopecia.Citation10,Citation11
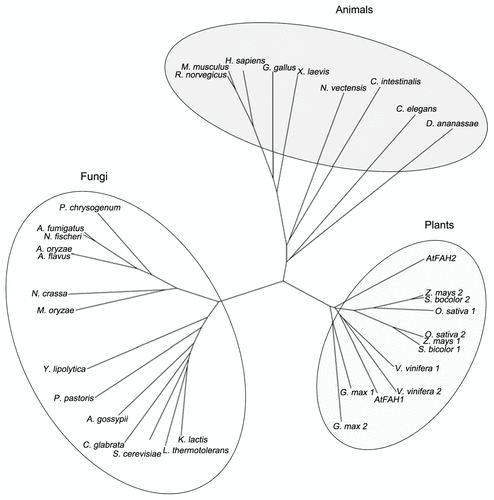
Recently, we identified two FAH homologs in Arabidopsis (Arabidopsis thaliana) (AtFAH1 and AtFAH2) that possessed the activity of fatty acid 2-hydroxylase,Citation12,Citation13 whereas S. cerevisiae and mammals have only a single gene as reported previously.Citation7,Citation8 In addition, neither AtFAH1 nor AtFAH2 possessed any Cb5-like domains and instead, both AtFAHs interacted with one of the ER-localized Cb5s (AtCb5-B) for activation. Moreover, AtFAH1 and AtFAH2 were used differently in fatty acid 2-hydroxylation and physiological functions. These differences suggest that plant FAHs may be quite different from their animal and fungus counterparts. We then searched for other FAH homologs in 20 animals, 31 fungi and 10 plants, and compared them based on their amino acid sequences. As shown in the phylogenetic tree created by representative ones, FAHs were apparently divided among plants, animals and fungi (). Interestingly, only one FAH homolog existed in all animals and fungi we searched in this study, whereas there were each two FAHs in plants such as Oryza sativa, Zea mays, Glycine max, Sorghum bicolor and Vitis vinifera, as well as Arabidopsis. This finding shows the peculiar character of plant FAHs in the number of FAHs. In plants, most of the complex sphingolipids contain a variety of 2-HFAs.Citation2 For example, there are 13 kinds of 2-HFAs from 16h:0 to 26h:1 and over 90% of glucosylceramides contain 2-HFAs.Citation14 In contrast, many fungi such as S. cerevisiae have only 26h:0, whereas complex sphingolipids in mammals contain only 50% 2-HFAs in limited tissues such as the nervous system and epidermal cells, although mammals have a diverse range of 2-HFAs from 16h:0 to 26h:0.Citation8,Citation15-Citation17 In addition, double mutant of AtFAH1 and AtFAH2 cannot be obtained until now, whereas the mammalian diseases by lack of FA2H were limited in the specific tissues and no serious phenotypes were reported in yeast FAH1 mutants. Therefore, it may be possible to speculate that plant FAHs have some plant-specific biological roles. On the other hand, the clade in plant FAHs was not unambiguously divided, suggesting that the function of FAHs was different even among plants. Moreover, it is unknown even whether multiple FAHs in individual plant species have different functions like Arabidopsis ones. Because the contents of 2-HFAs largely vary among plant species, it is interesting to investigate functional differentiation in each plant FAH to improve understanding of plant FAHs.
Figure 1. Phylogenetic relationship of FAH. A phylogenetic tree of FAH proteins derived from plants, animals and fungi was constructed by the CLUSTAL W program. Protein sequences were obtained from the National Center for Biotechnology Information. AtFAH1, At2g34770; AtFAH2, At4g20870; Oryza sativa 1, Os12g0628400; Oryza sativa 2, Os03g0780800; Glycine max 1, ACU24407; Glycine max 2, ACU24572; Sorghum bicolor 1, XP_002466355; Sorghum bicolor 2, XP_002442581; Vitis vinifera 1, XP_002268031; Vitis vinifera 2, XP_002267991; Homo sapiens, NP_077282; Mus musculus, NP_835187; Rattus norvegicus, NP_001129055; Gallus gallus, XP_414053; Xenopus laevis, NP_001082707; Nematostella vectensis, XP_001639353; Ciona intestinalis, XP_002126665; Caenorhabditis elegans, NP_492678; Drosophila ananassae, XP_001959617; Kluyveromyces lactis, XP_453142; Lachancea thermotolerans, XP_002552404; Saccharomyces cerevisiae, NP_013999; Candida glabrata, XP_446117; Ashbya gossypii, NP_982359; Pichia pastoris, XP_002492764; Yarrowia lipolytica, XP_505342; Magnaporthe oryzaeitalic>, XP_361446; Neosartorya fischeri, XP_955879; Aspergillus flavus, XP_002373274; Aspergillus oryzae, XP_001818088; Neosartorya fischeri, XP_001264167; Aspergillus fumigatus, XP_752948; Penicillium chrysogenum, XP_002561746.
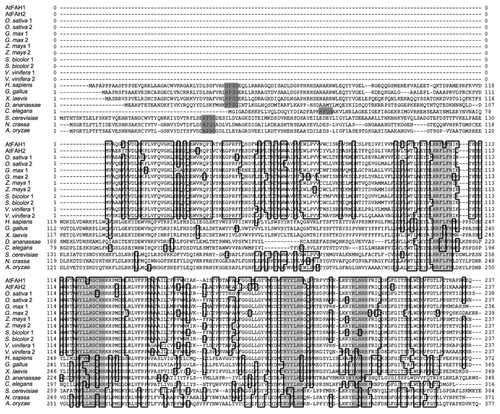
Next, we compared the amino acid sequences of FAHs among plants, animals and fungi. Oryza sativa, Zea mays, Glycine max, Sorghum bicolor and Vitis vinifera FAHs completely contained four histidine motifs similar to those of animal and fungus FAHs, suggesting that every plant FAH homolog may have the activity of fatty acid 2-hydroxylase like the Arabidopsis counterparts (, light gray). However, none of the plant FAHs had any Cb5-like domains, whereas all animal and fungus FAH homologs commonly conserved the domain in the N-terminus (, dark gray). Hence, it is possible that other plant FAHs also interact with Cb5 similar to the Arabidopsis ones and that the interaction leads to their activation.
Figure 2. Comparison of deduced amino acid sequences of FAH. Amino acid sequences were derived from Arabidopsis thaliana (AtFAH1, AtFAH2), Oryza sativa, Glycine max, Zea mays, Sorghum bicolor, Vitis vinifera, Homo sapiens, Gallus gallus, Xenopus laevis, Drosophila ananassae, Caenorhabditis elegans, Saccharomyces cerevisiae, Neurospora crassa and Aspergillus oryzae. Dark gray indicates HPGG, which is one of the most important motifs in the Cb5 protein family, and light gray indicates the histidine motif.
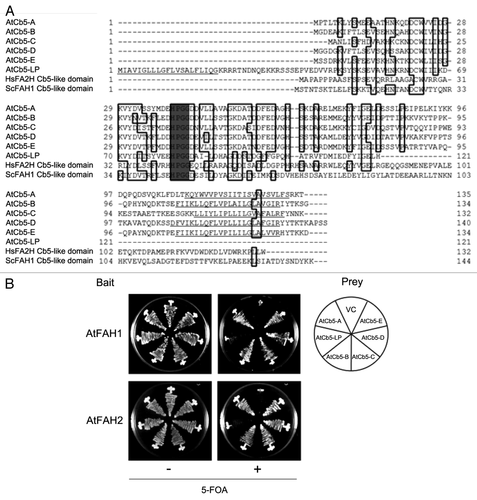
In plants, there are multiple ER-localized Cb5s, such as four in Arabidopsis (AtCb5-B, -C, -D and –E) and seven in rice, whereas mammals and yeast have only one ER-type Cb5.Citation12,Citation18 Thus, it is possible to speculate that the existence of a number of ER-type Cb5s complements the supply of electrons to FAHs without any Cb5-like domains in plants. When the amino acid sequences of AtCb5 family proteins including AtCb5-A (non-ER type) and AtCb5-like protein (AtCb5-LP, unknown function) were compared with those of Cb5-like domains of H. sapience FA2H (HsFA2H) and S. cerevisiae FAH1 (ScFAH1) proteins, all AtCb5 family proteins possessed crucial amino acids and were similar to Cb5-like domain of HsFA2H and ScFAH1, suggesting the possibility that every AtCb5 family protein supplies electrons to AtFAHs (). Then, we tested the interaction between AtFAHs and AtCb5 family proteins by using a split-ubiquitin yeast two-hybrid system.Citation12 In this experiment, only yeast cells containing the combination of interactors can survive on the medium with 5-fluoroorotic acid (5-FOA), because uracil expressed by the URA3 gene in the bait vector is disrupted by the effect of full-length ubiquitin.Citation19 When pMet-AtFAH and NuI-AtCb5s-myc were used as bait and prey constructs, respectively, both AtFAH1 and AtFAH2 interacted with AtCb5-A, -B, -C, -D and –E, although they did not interact with AtCb5-LP (). AtCb5-LP contains the motifs important for the Cb5 function, but it has a transmembrane domain in its N-terminus, which generally exists in the C-terminus of Cb5 (). For this reason, neither AtFAH1 nor AtFAH2 interacted with it. In addition, non-ER-type Cb5 in plants (AtCb5-A in this study) was found to be mislocalized in the ER of yeast because yeast has only the ER-type of Cb5.Citation20 This result indicates that both AtFAHs interact with all ER-localizing AtCb5s.
Figure 3. Interactions of AtFAHs with AtCb5 family proteins. (A) Comparison of deduced amino acid sequences among AtCb5 family proteins (AtCb5-A, -B, -C, -D, -E and -LP), and Cb5-like domains of human FA2H (HsFA2H) and yeast FAH1 (ScFAH1). Gray indicates HPGG. Possible transmembrane domains predicted by SOSUI WWW Server are underlined. (B) Interactions between AtFAHs and AtCb5 family proteins were detected by the suY2H system. Yeast cells containing bait and prey constructs were tested for their binding on minimum medium containing 5-fluoroorotic acid (5-FOA). The results indicate the growth of each line after 2 d (without 5-FOA) and 3 d (with 5-FOA) of incubation at 30°C. VC indicates the combination of vector controls.
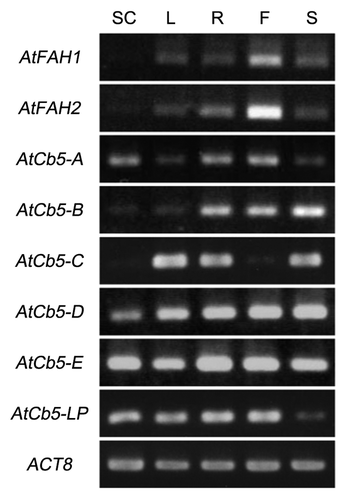
In addition, we checked the expression pattern of AtFAHs and AtCb5s in various tissues of Arabidopsis. AtFAH1 and AtFAH2 were expressed in all tissues, especially strongly in flowers, whereas little expression was detected in suspension cells (). On the other hand, in ER-type AtCb5s, AtCb5-B and AtCb5-C showed different expression patterns dependent on tissues, whereas AtCb5-D and AtCb5-E were highly expressed in all tissues (). However, none of the ER-type AtCb5s were expressed in the same way as AtFAH1 and AtFAH2. Together with the interaction data, this result suggests the possibility that AtFAH1 and AtFAH2 can use all ER-type AtCb5s, and that they use different AtCb5s in a tissue-dependent manner. In our pervious report, the activation of AtFAH1 and AtFAH2 by the direct interaction with AtCb5-B was demonstrated.Citation13 It is necessary to examine whether AtCb5-C, AtCb5-D and AtCb5-E also activate AtFAH1 and AtFAH2. Moreover, the result that both AtFAH1 and AtFAH2 were highly expressed in flowers may support the possibility that 2-HFAs play an important role in flowers. It would be interesting to further explore plant-specific physiological functions in various plant species.
Acknowledgments
Plasmids and strains used for the split-ubiquitin yeast two-hybrid system were generously provided by Dr. Ralph Panstruga (Max-Planck Institute, Saabruecken, Germany) and Dr. Imer E. Somssich (Max-Planck Institute). This research was supported by a Research Fellowship for Young Scientists of the Japan Society for the Promotion of Science (JSPS) (to M.N.), a grant in-aid to M.N. for Scientific Research for Plant Graduate Students from Nara Institute Science and Technology, a grant by the Japan Society for the Promotion of Science (JSPS) through the “Funding Program for Next Generation World-Leading Researchers (NEXT Program)” and a grant from the Ministry of Agriculture, Forestry, and Fishery, Japan (IPG0014).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Dunn TM, Lynch DV, Michaelson LV, Napier JA. A post-genomic approach to understanding sphingolipid metabolism in Arabidopsis thaliana.. Ann Bot 2004; 93:483 - 97; 10.1093/aob/mch071; PMID: 15037448
- Pata MO, Hannun YA, Ng CKY. Plant sphingolipids: decoding the enigma of the Sphinx. New Phytol 2010; 185:611 - 30; 10.1111/j.1469-8137.2009.03123.x; PMID: 20028469
- Hama H. Fatty acid 2-Hydroxylation in mammalian sphingolipid biology. Biochim Biophys Acta 2010; 1801:405 - 14; 10.1016/j.bbalip.2009.12.004; PMID: 20026285
- Pascher I, Sundell S. Molecular arrangements in sphingolipids, the crystal structure of cerebroside. Chem Phys Lipids 1977; 20:175 - 91; 10.1016/0009-3084(77)90033-0
- Löfgren H, Pascher I. Molecular arrangements of sphingolipids. The monolayer behaviour of ceramides. Chem Phys Lipids 1977; 20:273 - 84; 10.1016/0009-3084(77)90068-8; PMID: 597969
- Boggs JM, Koshy KM, Rangaraj G. Photolabeling of myelin basic protein in lipid vesicles with the hydrophobic reagent 3-(trifluoromethyl)-3-(m-[125I]iodophenyl)diazirine. Biochim Biophys Acta 1988; 938:361 - 72; 10.1016/0005-2736(88)90134-4; PMID: 3349071
- Mitchell AG, Martin CE. Fah1p, a Saccharomyces cerevisiae cytochrome b5 fusion protein, and its Arabidopsis thaliana homolog that lacks the cytochrome b5 domain both function in the alpha-hydroxylation of sphingolipid-associated very long chain fatty acids. J Biol Chem 1997; 272:28281 - 8; 10.1074/jbc.272.45.28281; PMID: 9353282
- Alderson NL, Rembiesa BM, Walla MD, Bielawska A, Bielawski J, Hama H. The human FA2H gene encodes a fatty acid 2-hydroxylase. J Biol Chem 2004; 279:48562 - 8; 10.1074/jbc.M406649200; PMID: 15337768
- Eckhardt M, Yaghootfam A, Fewou SN, Zöller I, Gieselmann V. A mammalian fatty acid hydroxylase responsible for the formation of alpha-hydroxylated galactosylceramide in myelin. Biochem J 2005; 388:245 - 54; 10.1042/BJ20041451; PMID: 15658937
- Schneider SA, Bhatia KP. Three faces of the same gene: FA2H links neurodegeneration with brain iron accumulation, leukodystrophies, and hereditary spastic paraplegias. Ann Neurol 2010; 68:575 - 7; 10.1002/ana.22211; PMID: 21031573
- Maier H, Meixner M, Hartmann D, Sandhoff R, Wang-Eckhardt L, Zöller I, et al. Normal fur development and sebum production depends on fatty acid 2-hydroxylase expression in sebaceous glands. J Biol Chem 2011; 286:25922 - 34; 10.1074/jbc.M111.231977; PMID: 21628453
- Nagano M, Ihara-Ohori Y, Imai H, Inada N, Fujimoto M, Tsutsumi N, et al. Functional association of cell death suppressor, Arabidopsis Bax inhibitor-1, with fatty acid 2-hydroxylation through cytochrome b₅. Plant J 2009; 58:122 - 34; 10.1111/j.1365-313X.2008.03765.x; PMID: 19054355
- Nagano M, Takahara K, Fujimoto M, Tsutsumi N, Uchimiya H, Kawai-Yamada M. Arabidopsis sphingolipid fatty acid 2-hydroxylases (AtFAH1 and AtFAH2) are functionally differentiated in fatty acid 2-hydroxylation and stress responses. Plant Physiol 2012; 159:1138 - 48; 10.1104/pp.112.199547; PMID: 22635113
- Imai H, Yamamoto K, Shibahara A, Miyatani S, Nakayama T. Determining double-bond positions in monoenoic 2-hydroxy fatty acids of glucosylceramides by gas chromatography-mass spectrometry. Lipids 2000; 35:233 - 6; 10.1007/BF02664774; PMID: 10757555
- Oh CS, Toke DA, Mandala S, Martin CE. ELO2 and ELO3, homologues of the Saccharomyces cerevisiae ELO1 gene, function in fatty acid elongation and are required for sphingolipid formation. J Biol Chem 1997; 272:17376 - 84; 10.1074/jbc.272.28.17376; PMID: 9211877
- Uchida Y, Hama H, Alderson NL, Douangpanya S, Wang Y, Crumrine DA, et al. Fatty acid 2-hydroxylase, encoded by FA2H, accounts for differentiation-associated increase in 2-OH ceramides during keratinocyte differentiation. J Biol Chem 2007; 282:13211 - 9; 10.1074/jbc.M611562200; PMID: 17355976
- Maldonado EN, Alderson NL, Monje PV, Wood PM, Hama H. FA2H is responsible for the formation of 2-hydroxy galactolipids in peripheral nervous system myelin. J Lipid Res 2008; 49:153 - 61; 10.1194/jlr.M700400-JLR200; PMID: 17901466
- Maggio C, Barbante A, Ferro F, Frigerio L, Pedrazzini E. Intracellular sorting of the tail-anchored protein cytochrome b5 in plants: a comparative study using different isoforms from rabbit and Arabidopsis.. J Exp Bot 2007; 58:1365 - 79; 10.1093/jxb/erl303; PMID: 17322552
- Wittke S, Lewke N, Müller S, Johnsson N. Probing the molecular environment of membrane proteins in vivo. Mol Biol Cell 1999; 10:2519 - 30; PMID: 10436009
- Hwang YT, Pelitire SM, Henderson MP, Andrews DW, Dyer JM, Mullen RT. Novel targeting signals mediate the sorting of different isoforms of the tail-anchored membrane protein cytochrome b5 to either endoplasmic reticulum or mitochondria. Plant Cell 2004; 16:3002 - 19; 10.1105/tpc.104.026039; PMID: 15486098