Abstract
To optimize water use efficiency, plants regulate stomatal closure through a complex signaling process. Hydrogen peroxide (H2O2) is produced in response to several environmental stimuli, and has been identified as a key second messenger involved in the regulation of stomatal aperture. The Arabidopsis histidine kinase 5 (AHK5) has been shown to regulate stomatal closure in response to H2O2 and other stimuli that depend on H2O2. AHK5 is a member of the two-component system (TCS) in Arabidopsis. The plant TCS comprises three different protein types: the hybrid histidine kinases (HKs), the phosphotransfer proteins (HPs) and the response regulators (RRs). Here we determined TCS elements involved in H2O2- and ethylene-dependent stomatal closure downstream of AHK5. By yeast and in planta interaction assays and functional studies, AHP1, 2 and 5 as well as the response regulators ARR4 and ARR7 were identified acting downstream of AHK5 in the ethylene and H2O2 response pathways of guard cells. Furthermore, we demonstrate that aspartate phosphorylation of ARR4 is only required for the H2O2- but not for the ethylene-induced stomatal closure response. Our data suggest the presence of a complex TCS signaling network comprising of at least AHK5, several AHPs and response regulators, which modulate stomatal closure in response to H2O2 and ethylene.
Introduction
Stomata are openings in leaves that regulate gas exchange and water loss between the plant and the environment. Guard cells surrounding stomatal pores regulate stomatal movements via swelling and shrinking in response to changes in turgor pressure, depending on the stimulus. Guard cells have been extensively used to not only understand plant responses to drought stress via the hormone abscisic acid (ABA), but also to other abiotic (salinity, ozone, light/dark) and biotic stimuli (bacteria, fungi and pathogen-derived elicitors).Citation1,Citation2 The extreme robustness of guard cell signaling networks, cell autonomy and the advantage of having a quick physiological read-out (stomatal aperture) make guard cells an extremely attractive system to study signal transduction pathways. This is over-ridden by the overall importance of transpirational water loss through stomata being crucial for plant survival under a constantly changing environment.
The signal transduction network in guard cells has been extensively studied for ABA, and is nowhere being complete.Citation2 However this has been a driving force to understand mechanisms of signal perception and transduction in other situations, e.g., the hormone ethylene and bacteria.Citation2,Citation3 Inevitably, cross-talk does occur between signaling pathways in guard cells; for example, between ABA and ethylene, as well as ABA and jasmonic acid (JA) and bacterial elicitors.Citation1,Citation3,Citation4 One of the key elements common to signal transduction cross-talk is the reactive oxygen species (ROS) hydrogen peroxide (H2O2). H2O2 has been shown to act as a central node integrating plant responses to both abiotic and biotic stimuli.Citation5 The mechanisms by which plant cells perceive ROS and trigger various signaling pathways are slowly being unraveled, particularly in guard cells.Citation6
In an attempt to understand redox signaling, we have recently shown that the hybrid histidine kinase AHK5 functions in Arabidopsis guard cells to integrate endogenous and environmental cues via redox homeostasis.Citation7 Although protein phosphorylation is a key signal transduction component in ABA-induced stomatal closure,Citation8 there is not much evidence for the two-component system (TCS) functioning in guard cells. The TCS network in plants consists of hybrid histidine kinases (HKs), histidine-containing phosphotransfer proteins (HPs) and response regulators (RRs). Signal perception by a HK leads to a phosphorelay from the HK, via HPs to a RR, modulating gene expression and other cellular reactions. The RR family in Arabidopsis (ARR) is divided into three subgroups, the A-, B- and C-type ARRs, according to their protein structure. The type-A ARRs are quite small proteins with a short receiver domain that contains the phosphorylatable aspartate residue and a short C-terminal extension.Citation9 They are upregulated by cytokinin and, apart from their function as negative regulators of cytokinin signalingCitation10 they have also been identified in other processes such as red light signalingCitation11,Citation12 circadian clockCitation13,Citation14 lateral root formation, leaf senescenceCitation15 and cold stress.Citation16
The type-B response regulator proteins (type-B ARRs) also comprise a receiver domain along with an extended C-terminal output region.Citation9 They act as transcription factors that induce transcription of type-A ARRs upon cytokinin treatment.Citation17 Structurally and functionally, the type-C ARRs resemble type-A ARRs; however, unlike type-A ARRs, their expression does not depend on cytokinin.Citation18
Our recent data show that the Arabidopsis HK (AHK) ETR1 functions in ethylene and H2O2-regulated stomatal closureCitation19 and the HK AHK5 functions in H2O2, NO, darkness, bacteria- and ethylene-induced stomatal closure.Citation7 Intriguingly, both these HKs appear not to be required for the ABA signal response pathway in guard cells.Citation7,Citation19
AHK5 is a unique HK protein in the Arabidopsis TCS family. It is the only HK that was predicted to have a cytosolic location; yet recent experiments showed that it is cytosolic and also associates with the plasma membrane.Citation7 This intracellular distribution pattern would make it unique in its ability to integrate various signals, both from within and outside the cell.Citation7 As AHK5 is a canonical HK, it is assumed to function within the TCS network. We therefore aimed to investigate the two-component elements in this signal transductome and how the interactors may function in the stomatal closure response. We show that AHK5 interacts with the Arabidopsis HP (AHP) proteins AHP1, AHP2 and AHP5 in vivo and that Arabidopsis plants carrying loss-of-function alleles of the AHP genes have defects in stomatal closure in response to ethylene and H2O2. Downstream of the AHK5-interacting AHPs, we identified ARR4 and ARR7 as RRs involved in the regulation of stomatal closure. In addition, the phosphorylation of ARR4 appears to be required for at least some of AHK5-dependent signal transduction to occur.
Results
Identification of AHPs that associate with the AHK5 protein
AHK5 is a canonical hybrid histidine kinase and, thus, proposed to be the first element in a phosphorelay process that also consists of AHPs and ARRs. This opens the question of whether AHK5 function in stomatal closure rests upon a classical TCS pathway. Until now, no TCS element has been described acting downstream of AHK5. The TCS elements acting immediately downstream of hybrid AHKs are the AHPs. To elucidate which of the 5 canonical AHPs (AHP1, 2, 3, 4, 5) may act down-stream of AHK5, we first performed AHK5-AHP interaction studies using the yeast two-hybrid system (Y2H). As shown in , AHK5 interacted with AHPs 1, 2, 3 and 5 but not with AHP4 in Y2H, although AHP4 was expressed in the yeast cells ().
Figure 1. AHK5 interacts with a set of AHP proteins in yeast-two-hybrid assays. (A) Yeast-two-hybrid assay with yeast cells co-expressing BD::AHK5 (AHK5 protein fused to the BD domain of the Gal4 DNA) and the indicated AD::AHP (AHP1 to 5 proteins fused to the AD domain of the Gal4 DNA) fusion proteins. Yeast cells were incubated for 4 d at 28°C on either vector selective (L-, W-) or interaction selective media (L-, W-, Ade-) (B) Western-blot analysis using crude extracts from transformed yeast cells co-expressing the indicated AD::AHP (AHP1 to 5) and BD::AHK5 fusion proteins. The AD::AHP (upper panel) and the BD::AHK5 (lower panel) fusions were detected with a HA- and c-myc-specific antibody respectively.
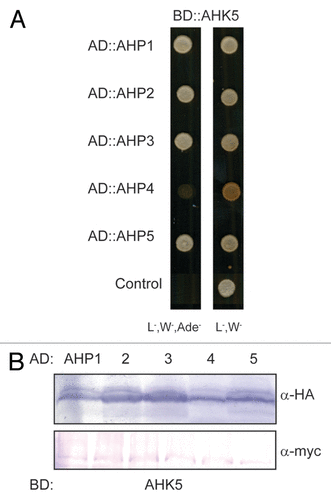
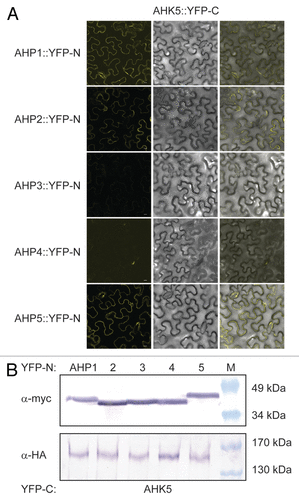
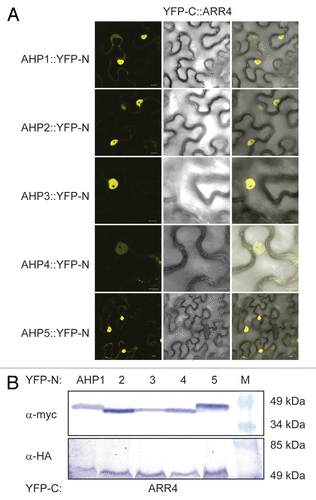
To substantiate the yeast results with an independent technique, we also performed in planta protein-protein interaction studies based on a bimolecular fluorescence complementation (BiFC) assay in transiently transformed tobacco (Nicotiana benthamiana) leaf epidermal cells.Citation20,Citation21 Although AHK5 and the different AHP proteins co-accumulated to a similar level in the independently co-transformed tobacco leaves (), BiFC signals were only observed in the case where AHK5 was co-expressed with AHP1, 2 and 5 (). This indicates that AHP1, 2 and 5 are the specific interaction partners of AHK5 in planta and that the interaction of AHK5 with AHP3 is probably a yeast artifact. The divergence of the yeast and in planta results could be due to the fact that yeast represents a heterologous system lacking plant components which are required to achieve a more specific interaction pattern in the TCS network.
Figure 2. AHK5 interacts with a set of AHP proteins in a Bimolecular Fluorescence Complementation Assay (BiFC). (A) Confocal images of epidermal tobacco leaf cells (Nicotiana benthamiana) co-expressing the indicated YFP-N and YFP-C fusion proteins. The left panels show the fluorescence signal, the middle panels the bright field images of identical cells and the right panels the overlay of both. YFP-N, N-terminal YFP fragment fused to the different AHP proteins; YFP-C, C-terminal YFP fragment fused to the AHK5 protein. The bars represent 10 μm. (B) Western-blot analysis using crude protein extracts from transiently transformed tobacco leaves analyzed for BiFC before extraction. The AHP::YFP-N (upper panel) and the AHK5::YFP-C (lower panel) fusions were detected with a c-myc and HA-specific antibody respectively. M, protein marker.
Behavior of ahp mutants to the AHK5-dependent ethylene and H2O2-induced stomatal closure response
The data above suggest a putative TCS signaling pathway that would initiate with AHK5 and carry on with AHP1, 2 and 5. As mentioned above, AHK5 is involved in H2O2-induced as well as in the ethylene-induced stomatal closure response in Arabidopsis. To test if AHK5-AHPs interactions are of physiological relevance for the ethylene-induced stomatal closure response, we analyzed the phenotypic behavior of ahp1, 2 and 5 mutantsCitation22 in the presence of ethephon, an ethylene-releasing agent. We also included the remaining AHP mutants, namely ahp3 and ahp4,Citation22 whose wild type proteins did not interact with AHK5 in plant cells, to analyze the entire family of AHP proteins in Arabidopsis. In ahk5 mutants, the ethylene response is impaired, resulting in plants that, opposite to the wild type, do not close their stomata in response to the hormone (Fig. S1).Citation7 The stomatal aperture response of ahp mutants treated with ethephon relative to their corresponding mock-treated controls (100%), have been plotted in . Interestingly, ahp1, ahp2 and ahp3 single mutants displayed a significant difference in their stomatal response to ethephon, compared with WS and Col-0 ethephon treated stomata (). In contrast, ahp4 and ahp5 mutants displayed a wild type response to ethephon, despite the control apertures for ahp5 being considerably smaller than wild type Col-0 (Fig. S2). These data suggest a function for AHPs 1, 2 and 3 alone for ethylene-induced stomatal closure.
Figure 3. The ethylene and H2O2-induced stomatal response pathway downstream of AHK5. (A) Stomatal aperture in leaves of wild type (WS, Col-0) and different ahp Arabidopsis mutants. The leaves were exposed to 100 µM ethephon for 2h. Data are expressed as stomatal aperture response relative to mock-treated controls. Data are derived from measuring the apertures of at least 60 stomata from 3 independent experiments. * = Statistical difference (p-value ≤ 0.001 via Student’s t-test) compared with the wild type plants. (B) Stomatal aperture in leaves of wild type (WS, Col-0) and different ahp Arabidopsis mutants. The leaves were exposed to 100 µM H2O2 for 2h. Data are expressed as stomatal aperture relative to mock-treated controls. Data are derived from measuring the apertures of at least 60 guard cells from 3 independent experiments. * = Statistical difference (p-value ≤ 0.001 via Student’s t-test) compared with the wild type plants.
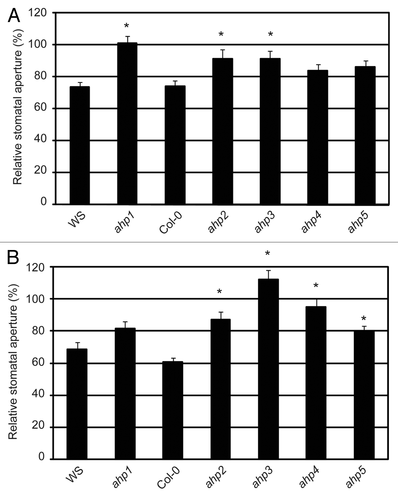
Given that ethylene and H2O2 are linked to the AHK5-dependent guard cell signaling response, the role of the AHK5-interacting AHPs in the H2O2-dependent stomata closure pathway was also determined using the above mutants. shows that only ahp1 stomata responded to H2O2 in a manner similar to wild type WS guard cells. However, compared with wild type Col-0 response to H2O2, the ahp2, ahp4 and ahp5 mutants responded significantly less to H2O2 treatment. It must be noted that although ahp5 stomata did close to H2O2 (Fig. S2) the response was significantly less compared with Col-0 (). In contrast, stomata of the ahp3 mutant were slightly, but significantly, more open with H2O2. Therefore, of the AHPs that interact with AHK5 in planta, AHP1 appears to only regulate ethylene-induced stomatal closure. AHP5 functions only in the H2O2-induced stomatal closure pathway, while AHP2 is essential for both the ethylene and H2O2 response. Interestingly, although AHP3 and AHP4 do not appear to interact with AHK5, AHP3 still regulates both ethylene and H2O2-induced stomatal closure, while AHP4 regulates H2O2-induced stomatal closure alone ().
Identification of putative ARRs that could be involved in the AHK5 two-component system
After elucidating the AHP components that were involved in the stomatal response, the final TCS output element, in this case the specific ARR, responsible to conclude the TCS pathway remained to be established. There are a total of 32 ARRs that have been described in ArabidopsisCitation23 and of these, ARR2 was shown to be functioning as a positive element downstream of ETR1 in the ethylene and H2O2 signal transduction pathway in guard cells.Citation3 To be downstream of the AHK5-AHP signaling pathway, the ARR must be able to interact and function with at least AHP1, AHP2 or AHP5. By Y2H assays it has been shown that 10 ARRs including members of all three A-, B- and C-type subgroups interacted in yeast with AHP1, 2 and 5, namely ARR1, 2, 3, 4, 5, 7, 8, 9, 14, 16 and 22.Citation24,Citation25 From these, only the A-type ARR4 and ARR7, the B-type ARR1 and ARR2 and the C-type ARR22 are significantly expressed in stomatal guard cells.Citation26,Citation27 Here we focus on the function of the A-type ARRs in the ethylene and H2O2 signaling pathways downstream of AHK5 in guard cells. We recently demonstrated that the A-type response regulator ARR4 is not only able to interact with AHP1 in the Y2H system but also accepts phosphoryl residues from AHP1 in a plant cell-derived in vitro phosphorelay system.Citation11 This makes ARR4 a promising and most obvious candidate downstream of the AHK5-dependent phosphorelay.
In order to verify that ARR4 actually interacts with those AHPs which also associated with AHK5 in plant cells, we repeated the Y2H interaction assays and in addition, performed in planta BiFC interaction studies in tobacco. We could confirm that ARR4 interacts with AHP1, 2, 3 and 5, and excluded AHP4 as an interaction partner in yeast (). Furthermore, the results of the in planta BiFC interaction studies corroborated the yeast interaction data: ARR4 interacted in planta with AHP1, 2, 3 and 5 but not with AHP4 ().
Figure 4. ARR4 interacts with a set of AHP proteins in yeast-two-hybrid assays. (A) Yeast-two-hybrid assay with yeast cells co-expressing BD::ARR4 or BD::ARR7 (ARR4 or ARR7 protein fused to the BD domain of the Gal4 DNA) and the indicated AD::AHP (AHP1 to 5 proteins fused to the AD domain of the Gal4 DNA) fusion proteins. They were incubated for 4 d at 28°C on either vector selective (L-, W-) or interaction selective media (L-, W-, Ade-) (B) Western-blot analysis using crude extracts from transformed yeast cells co-expressing the indicated AD::AHP (AHP1 to 5) and BD::ARR4 (B) or BD::ARR7 (C) fusion proteins. The AD::AHP (upper panel) and the BD::ARR4 or BD::ARR7 (lower panels) fusions were detected with a HA- and c-myc-specific antibody respectively.
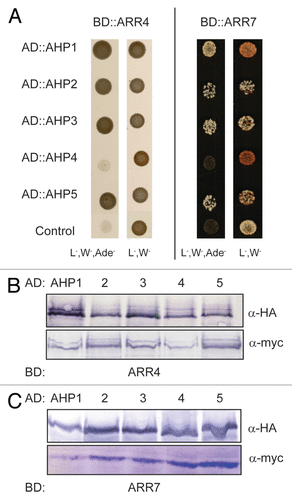
Figure 5. ARR4 interacts with a set of AHP proteins in a Bimolecular Fluorescence Complementation Assay (BiFC). (A) Confocal images of epidermal tobacco leaf cells (Nicotiana benthamiana) co-expressing the indicated YFP-N and YFP-C fusion proteins. The left panels show the fluorescence signal, the middle panels the bright field images of identical cells and the right panels the overlay of both. YFP-N, N-terminal YFP fragment fused to the different AHP proteins; YFP-C, C-terminal YFP fragment fused to the ARR4 protein. The bars represent 10 μm. (B) Western-blot analysis using crude extracts from transiently transformed tobacco cells co-expressing the indicated AHP::YFP-N (AHP1 to 5) and YFP-C::ARR4 fusion proteins. The AHP::YFP-N (upper panel) and the YFP-C::ARR4 (lower panel) fusions were detected with a c-myc- and HA-specific antibody respectively. M, protein marker.
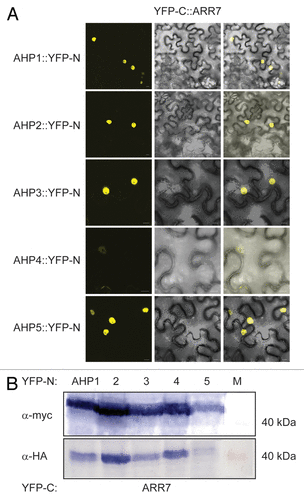
We further extended our studies to include the other type A ARR, ARR7 which is also expressed in guard cells. Both Y2H and BiFC interaction studies in planta concluded that like ARR4, ARR7 interacts with AHP1, 2, 3 and 5 but not AHP4 (Fig. Four and 6). Together with the above AHK5-AHP data, the possibility therefore arises that AHK5, AHP1, AHP2, AHP5 and ARR4/ARR7 comprise a TCS signaling pathway, which might regulate guard cell activity in response to H2O2 and ethylene.
ARR4 and ARR7 differentially contribute to the ethylene- and H2O2-induced stomatal closure response
To test the significance of ARR4 in the ethylene-induced stomatal closure response, we tested the response of the arr4 T-DNA insertion mutantCitation28 to ethylene. In contrast to wild type Col-0 plants, arr4 stomata did not close when treated with ethephon (). Taking into account the functional redundancy described for A-type response regulators in processes such as cytokinin signaling,Citation28 it is likely that other response regulators apart from ARR4 intervene in the AHK5-induced stomatal closure response to both ethylene and H2O2. ARR7 together with ARR4 are the only A-type ARRs that interact with AHP1 and AHP2Citation24 and are expressed in guard cells,Citation27 thus ARR7 might also act as an element downstream of AHK5 in this response. We, therefore, included the arr7 mutant in our study.Citation29 Stomata of the arr7 mutant did not close as much as wild-type Col-0 upon ethephon treatment ( and Fig. S3). Therefore both ARR4 and ARR7 appear to regulate ethylene-induced stomatal closure.
Figure 7. The ethylene and H2O2-induced stomatal response pathway downstream of AHK5 involves ARR4 and ARR7. (A) Stomatal aperture in leaves of wild type (Col-0, WS), arr4, arr7 and ARR4D95N (non-phosphorylatable ARR4 overexpressor) Arabidopsis plants. The leaves were exposed to 100 µM ethephon for 2h. Data are expressed as stomatal aperture response relative to mock-treated controls. Data are derived from measuring the apertures of at least 60 stomata from 3 independent experiments. * = Statistical difference (p-value ≤ 0.001 via Student’s t-test) compared with the wild type plants. (B) Stomatal aperture in leaves of wild type (Col-0, WS), arr4, arr7 and ARR4D95N (non-phosphorylatable ARR4 overexpressor) Arabidopsis plants. The leaves were exposed to 100 µM H2O2 for 2h. Data are expressed as stomatal aperture response relative to mock-treated controls. Data are derived from measuring the apertures of at least 60 stomata from 3 independent experiments. * = Statistical difference (p-value ≤ 0.001 via Student’s t-test) compared with wild type plants.
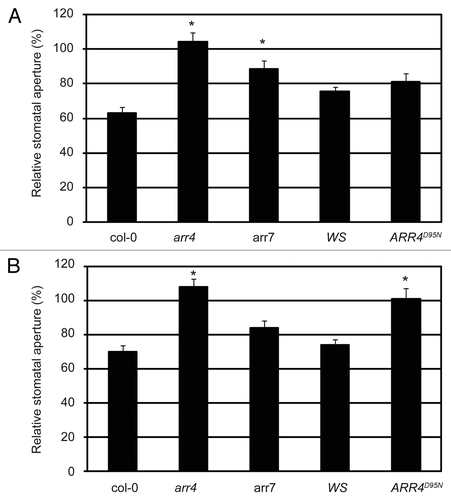
Figure 6. ARR7 interacts with a set of AHP proteins in a Bimolecular Fluorescence Complementation Assay (BiFC). (A) Confocal images of epidermal tobacco leaf cells (Nicotiana benthamiana) co-expressing the indicated YFP-N and YFP-C fusion proteins. The left panels show the fluorescence signal, the middle panels the bright field images of identical cells and the right panels the overlay of both. YFP-N, N-terminal YFP fragment fused to the different AHP proteins; YFP-C, C-terminal YFP fragment fused to the ARR7 protein. The bars represent 10 μm. (B) Western-blot analysis using crude extracts from transiently transformed tobacco cells co-expressing the indicated AHP::YFP-N (AHP1 to 5) and YFP-C::ARR7 fusion proteins. The AHP::YFP-N (upper panel) and the YFP-C::ARR7 (lower panel) fusions were detected with a c-myc- and HA-specific antibody respectively. M, protein marker.
TCS signaling mechanisms are based on a phosphorelay that transfers the signal from the hybrid histidine kinase, over the phosphotransfer proteins, to the response regulators. It was, therefore, important to determine whether the phosphorylation of ARR4 is required for ethylene-induced stomatal closure and to investigate whether the response was affected in a transgenic line, which ectopically expresses a non-phosphorylatable, loss-of-function version of ARR4 (ARR4D95N).Citation11 Surprisingly, the ARR4D95N-expressing line behaved as wild type WS plants, closing its stomata normally upon ethylene treatment (). This suggests that, although ARR4 is involved in the signaling pathway, its activation through aspartate phosphorylation is not required for the ethylene response.
To test the function of ARR4 and ARR7 in the H2O2 signaling pathway leading to stomatal closure, leaves from the above mutants were treated with exogenous H2O2 and their stomatal closure response was measured. The arr4 mutant clearly displayed insensitivity to H2O2 treatment, whereas arr7 showed stomatal closure (), not significantly different from that of wild type Col-0 (Fig. S3). Interestingly, the transgenic line which ectopically expresses non-phosphorylatable ARR4D95N, showed a significantly impaired stomatal closure response upon H2O2 treatment when compared with wild type Ws ( and Fig. S3), suggesting that phosphorylation of ARR4 is necessary for H2O2-induced stomatal closure.
In conclusion, our biochemical, genetic and physiological data show that AHK5 interacts with AHPs 1, 2, 5 and ARR4/ARR7, to regulate stomatal closure responses and that ARR4 phosphorylation is only required in the H2O2- but not the ethylene-induced stomatal closure response.
Discussion
AHK5 is a unique histidine kinase, in that it is the only predicted cytosolic HK known in Arabidopsis. In previous work we showed that AHK5 is not only located in the cytosol, but is also associated with the plasma membrane.Citation7 Our previous genetic and physiological analyses showed that AHK5 acts to integrate endogenous and external cues to regulate stomatal closureCitation7 but still the mechanism of AHK5 action in guard cells remains to be elucidated. Here we demonstrate that AHK5 acts as part of a two-component system, physically interacting with the phosphotransfer proteins AHP1, AHP2 and AHP5. These AHPs subsequently interact with the response regulator proteins ARR4 and ARR7. We also show that functional AHP1, AHP2, AHP5 and ARR4/ARR7 proteins are required for stomatal closure response to either ethylene and/or H2O2.
Previously, numerous studies have used yeast two-hybrid assays and in vitro co-purification experiments with recombinant proteins to determine interaction of plant HKs with AHPs and AHPs with ARRs.Citation24,Citation30,Citation31 However, the functional relevance of these interactions needs to be verified in planta, in most cases. Our data here and elsewhereCitation32 show that the BiFC assay is reliable to demonstrate physical interaction between the two component system proteins. AHK5 has been shown to have histidine kinase activityCitation33 and shown to complement yeast SLN1 mutants.Citation34 Therefore it was necessary to show that AHK5 does indeed function in a two-component system – our results show for the first time that AHK5 interacts with AHP1,2 and 5 (but not AHPs 3 and 4) in planta.
To extend the biochemical interaction data to functional significance, we tested insertion mutants of these AHPs for their stomatal response to ethylene and H2O2, two different stimuli shown to affect the stomatal closure response of ahk5 (see Fig. S1).Citation7 For the ethylene response, it appears that AHP1 and AHP2 function are essential, but AHP5 function is not required. As expected, when the response to ethylene-induce stomatal closure was tested on ahp4 mutant, an AHP element that is not downstream of the AHK5 pathway, the response observed was similar to wild type plants and the ahp4 mutant closed its stomata. In contrast, ahp3 mutants displayed an impaired response to ethylene, although AHP3 does not interact with AHK5 and is therefore, not a direct downstream element of AHK5. This leads to the conclusion that, apart from AHK5, other AHKs (either ETR1 or other HKs) are also responsible for the ethylene-regulated stomatal closure response.
H2O2 is a key signaling molecule generated in response to multiple stimuli, and of relevance to this work, shown to be required for a stomatal closure response to ethylene.Citation3 Previously we showed that AHK5 regulates redox homeostasis in stomatal guard cells in response to ethylene and the bacterial elicitor flagellin.Citation7 Here we find that the signaling pathway for H2O2-induced stomatal closure via AHK5-interacting AHPs appears to be different to that observed for ethylene. In contrast to the ethylene response, AHP1 is clearly not involved in H2O2-induced closure. However, both AHP2 and AHP5 regulate H2O2-induced closure. The fact that ahp4 and ahp3 mutants do not display a wild type response to H2O2 treatment hints that, as we propose for ethylene signaling, other HKs different to AHK5 are involved in H2O2-induced stomatal closure responses.
Together the above complex observations suggest that there are different routes to the AHK5-dependent TCS, depending on the input stimulus. So while AHP2 appears to be overlapping in both signaling pathways, AHP1 appears to be unique to the ethylene-signaling arm alone while AHP5 is unique to the H2O2 branch of the pathway (). The AHP1-dependent ethylene pathway indicates that there is a H2O2-independent pathway operating downstream of ethylene in guard cells. Ongoing work in our laboratory indicates that nitric oxide (NO) is also generated following ethylene-stimulation of guard cells. It is possible that the NO pathway occurs independent of the H2O2 signaling branch, where AHP1 might fit in. Given the robustness of guard cell signaling networks, apart from the ETR1- and the AHK5-dependent route, other AHKs could also be involved in the ethylene and H2O2-induced stomatal closure response in Arabidopsis. From the physiological data, we can conclude that the AHP proteins downstream of these unknown AHKs are AHP3 in the ethylene and both AHP3 and AHP4 in the H2O2 response.
Figure 8. Hypothetical model of ethylene and H2O2-induced stomatal closure responses via AHK5. The H2O2 branch of the pathway (red arrows) occurs via AHK5- or ETR1-activated AHP2/AHP5 signaling which leads to the activation and phosphorylation of ARR4, and thus resulting in stomatal closure. The stimulus which generates H2O2 in guard cells and functions via ARR4 phosphorylation remains to be determined. Ethylene stimulates H2O2 production via ETR1/AHK5. Ethylene response occurs via AHK5-AHP1/AHP2-ARR4 pathway (blue arrows), independent of ARR4 phosphorylation or via ARR7. In the H2O2-independent branch (dashed lines), AHP1 or AHP3 might act via ARR2 which induces ARR4 transcription, followed by protein accumulation; ethylene signaling therefore requires ARR4 function for stomatal closure. Black arrow represents transcriptional control.
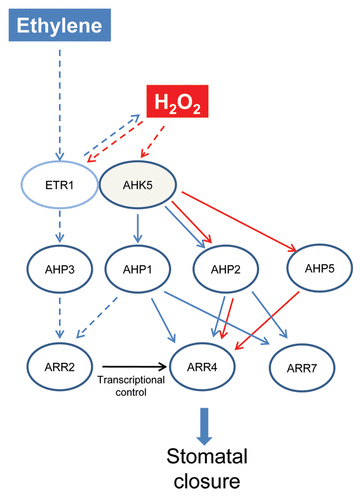
To identify the last component(s) of the AHK5-dependent TCS pathway, yeast interaction assays and BiFC experiments were performed. We demonstrated that of the various canonical AHPs, AHP1, 2, 3 and 5 interact with ARR4 and ARR7. ARR4 and ARR7 are A-type response regulators, having only a receiver domain and a very short output domain.Citation35 ARR4 has been shown to not only participate as a negative regulator in cytokinin signaling, but also as a positive regulator modulating the phytochrome B and other environmental signaling pathways.Citation11 ARR7 functions in cytokinin-dependent meristem function.Citation29 Here we provide functional evidence that ARR4 functions as a positive regulator in the ethylene- and H2O2-induced stomatal closure pathways, while ARR7 functions in the ethylene pathway alone. However, rather surprisingly, our genetic data demonstrate that phosphorylation of ARR4 is required only for the H2O2 response, but not for the ethylene response in stomata. This suggests that ARR4 may function via phosphorylation-independent interference with other proteinsCitation30 in the stomatal ethylene response pathway.
We postulate that one of these proteins might be the type-B RR ARR2. Both ethylene signaling in guard cells and a phosphorelay downstream of the ethylene receptor ETR1 involve the ARR2 protein.Citation3,Citation36 Furthermore, ETR1 and ARR2 have also been shown to interact with AHP1, AHP2 and AHP3 in vitro.Citation24,Citation30,Citation31,Citation37 Our data suggest that AHPs 1, 2 and 3 alone function in the ethylene stomatal closure pathway. Given that AHP2 is common to the H2O2 pathway this leaves either AHP1 or AHP3 to potentially act upstream of ARR2 in the ethylene pathway independent of H2O2 (). This would imply that upon ethylene perception by the ethylene receptor ETR1, a phosphorelay involving different AHPs could take place to activate the ARR2 response regulator. ARR2 encodes a transcriptional regulator known to activate A-type ARR genes.Citation18 ARR2 activation by ethylene could thus result in an enhanced expression of the A-type ARR4 followed by an enhanced ARR4 protein accumulation. ARR4 could then mediate the stomatal ethylene response in a phosphorylation-independent manner ()
On the other hand, the phosphorylation-dependent ARR4 pathway occurring via H2O2 must be occurring in response to other stimuli (such as bacterial elicitors) that generate H2O2; this remains to be determined. H2O2 signaling in guard cells seems to involve different routes. We established one that consists of AHK5, AHP2, AHP5, ARR4 and ARR7. However, which AHKs and ARRs participate in H2O2 signaling involving AHP3 and AHP4 needs to be clarified.
The complexity arising from these new data in the ethylene signaling pathway in guard cells suggest that cross-talk between the HKs AHK5 and ETR1 are occurring. It has been shown that AHK5 acts to inhibit the ETR1-dependent ethylene pathway leading to root growth inhibition.Citation33 The HK function of ETR1 appears to be required at least for the ethylene triple response and leaf development.Citation38 However in stomatal guard cells, the HK domain of ETR1 appeared to not be essential for H2O2-induced stomatal closure.Citation19 We speculate that the N-terminus of ETR1 may interact with the H2O2-activated AHK5 in the ethylene signaling pathway in guard cells.Citation7 Our data here support this hypothesis, in that the downstream components of AHK5 are essential for ethylene signaling. However, robustness of guard cell signaling dictates that there is always more than one signal transduction pathway triggered in response to a given stimulus. Therefore the alternative H2O2-independent pathway downstream of ethylene could also be independent of AHK5 ().
To summarize this data, we identified TCS downstream elements of the AHK5-dependent ethylene- and H2O2-induced stomatal closure response in Arabidopsis. We describe a novel ethylene-induced stomatal closure response that is independent of H2O2 and phosphorylation of a RR. Our data highlight new complexities in the ethylene signal transduction pathway which have not been described before. Stomatal guard cells have proven to be a valuable system to identify new components in signaling pathways. Nevertheless to understand the robustness of this system and unravel which of the key nodes is essential for ethylene signaling, mathematical modeling is needed alongside detailed quantification of various key steps leading to stomatal closure.
Materials and Methods
Plant growth conditions
Nicotiana benthamiana plants that were used for transient transformation of BiFC-constructs were cultivated in the greenhouse in a day/night cycle of 14h/10h. Temperature: day 25°C/night 19°C; 60% humidity. For growth of Arabidopsis plants, seeds were sown on Levington’s F2 compost + sand and maintained in controlled environment growth rooms for 4 weeks under short day conditions of 10 h light/14 h dark cycles with a light intensity of 120–150 µE m−2 s−1 at a temperature of 23°C and 55–65% relative humidity.
Stomatal bioassays
Leaves were floated for 2h under continuous illumination (120–150 µE m−2 s−1) in MES/KCl buffer (5mM KCl/10mM MES/50µM CaCl2, pH 6.15). Once the stomata were fully open, leaves were treated with either ethephon or H2O2 for further 2h. The leaves were subsequently homogenized individually in a Waring blender for 30 sec and the epidermal fragments collected on a 100 µm nylon mesh (SpectraMesh, BDH-Merck, UK). Stomatal apertures from epidermal fragments were then measured using a calibrated light microscope attached to an imaging system (Leica QWin software, Leica, UK). To calculate stomatal aperture relative values, first, both mock-treated (X) and treated (XT) stomatal aperture mean values were calculated for each genotype using the formula:
where n represents the number of stomata measured per line and Xi and XTi, the whole data set of stomatal aperture values per treatment. Finally, each genotype could be given a relative mean value (XR) calculated as follows: XR = XT/X.
In order to compare the t-tests of relative data, we did an estimation of the standard deviation (SD) of the relative stomatal apertures according to the Gaussian rules of error propagation. The relative SD (SXR) was calculated as follows with mean values X, XT and XR of data samples Xi, XTi and XRi and SD values SX, SXT and SXR respectively:
(SXR/XR)2 = (SXT/XT)2 + (SX/X)2
The different values for the data sample XRi, required to compare the t-tests of relative data, were calculated from each treated sample value (XTi) using the following formula: XRi = XTi /X.
Cloning strategy
All clones were constructed using Gateway™ technology (Invitrogen). The Entry clones were generated by BP-reaction, combining the genes of interest in pDONR207 (Invitrogen). All genes were amplified using cDNA preparations from Arabidopsis plant material. To avoid spontaneous mutations, the Entry clone of AHK5 was cloned and propagated using the E. coli strain CopyCutter™ (Epicenter).
For the BiFC assays, the AHP cDNAs were recombined via LR-reaction into pSPYNE-35S and AHK5 into pSPYCE-35S, which C-terminally express the N- and C-terminal YFP-fragments respectively.Citation21 The ARR4 cDNA was combined into pSpyCe-GW binary destination vector, which N-terminally expresses the C-terminal YFP-fragment.Citation39
Yeast-two-hybrid experiments were performed using the Matchmaker™ System (Clontech, USA). The BD-AHK5 and BD-ARR4 plasmids were constructed by LR-reaction using the corresponding Entry clones and the destination vector pGBKT7-DEST. To generate AD-AHP1–6, the destination vector pGADT7-DEST was used.
Yeast-two-hybrid analysis
The BD- and AD-destination clones were co-transformed into the yeast strain PJ69–4A, using lithium acetate/SS-DNA/PEG method.Citation40 After transformation, the yeast was grown on vector selective medium (CSM-L-, W-) and incubated for 3 d at 28°C. Subsequently, 5 independent yeast clones were pooled and grown in liquid vector selective medium to an OD600 of 1.0. To test protein interaction, 7.5µl of each culture were dropped on interaction selective medium (CSM-L-, W-, Ade-) and incubated for 2 d at 28°C and. As a control for testing the presence of the AD- and BD-fusion proteins, the yeast culture was also dropped on vector selective medium.
Infiltration of BiFC-constructs into Nicotiana benthamiana leaves
Transient transformation of Nicotiana benthamiana leaves using the Agrobacterium tumefaciens strain GV3101 pMP90, previously transformed with the desired BiFC-constructs, was performed as previously described.Citation20
Two to three days post infiltration, Nicotiana benthamiana epidermal leaf cells were analyzed for fluorescence (protein interaction) by confocal laser scanning microscopy using a Leica TCS SP2 confocal microscope (Leica Microsystems GmbH). The microscopy was performed as described previously.Citation25
Protein extraction, SDS-PAGE, western blot and immunodetection
Western Blot analysis was used to verify the correct expression of the desired fusion proteins in BiFC- and Y2H-assays. Leaf discs were excised from infiltrated tobacco plants and proteins were extracted using denaturing SDS sample buffer.Citation20 Total protein extract analyzed via SDS-PAGE and subsequent Western Blot and immunodetection using HA- and c-myc-specific antibodies (Roche, UK) was performed as described.Citation20 Protein extraction from transformed yeast cells was performed as done previously.Citation40
Additional material
Download Zip (189.8 KB)Acknowledgments
We thank J. Kieber, G.E. Schaller and J.U. Lohmann for arr4, arr7 and ahp seeds. This work was funded by DFG grants to K.H. (HA 2146/10–1, HA 2146/16–1), a DAAD PhD fellowship to M.V. and PhD fellowship of the University of Tübingen to J.W.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Schroeder JI, Kwak JM, Allen GJ. Guard cell abscisic acid signalling and engineering drought hardiness in plants. Nature 2001; 410:327 - 30; 10.1038/35066500; PMID: 11268200
- Underwood W, Melotto M, He SY. Role of plant stomata in bacterial invasion. Cell Microbiol 2007; 9:1621 - 9; 10.1111/j.1462-5822.2007.00938.x; PMID: 17419713
- Desikan R, Last K, Harrett-Williams R, Tagliavia C, Harter K, Hooley R, et al. Ethylene-induced stomatal closure in Arabidopsis occurs via AtrbohF-mediated hydrogen peroxide synthesis. Plant J 2006; 47:907 - 16; 10.1111/j.1365-313X.2006.02842.x; PMID: 16961732
- Suhita D, Raghavendra AS, Kwak JM, Vavasseur A. Cytoplasmic alkalization precedes reactive oxygen species production during methyl jasmonate- and abscisic acid-induced stomatal closure. Plant Physiol 2004; 134:1536 - 45; 10.1104/pp.103.032250; PMID: 15064385
- Desikan R, Cheung MK, Bright J, Henson D, Hancock JT, Neill SJ. ABA, hydrogen peroxide and nitric oxide signalling in stomatal guard cells. J Exp Bot 2004; 55:205 - 12; 10.1093/jxb/erh033; PMID: 14673026
- Pham J, Desikan R. ROS signalling in stomata. In: Reactive oxygen Species in Plant Signalling. Puppo A, del Rio L, Eds. Springer-Verlag 2009; pp55-71.
- Desikan R, Horák J, Chaban C, Mira-Rodado V, Witthöft J, Elgass K, et al. The histidine kinase AHK5 integrates endogenous and environmental signals in Arabidopsis guard cells. PLoS One 2008; 3:e2491; 10.1371/journal.pone.0002491; PMID: 18560512
- Schroeder JI, Allen GJ, Hugouvieux V, Kwak JM, Waner D. Guard Cell Signal Transduction. Annu Rev Plant Physiol Plant Mol Biol 2001; 52:627 - 58; 10.1146/annurev.arplant.52.1.627; PMID: 11337411
- Grefen C, Harter K. Plant two-component systems: principles, functions, complexity and cross talk. Planta 2004; 219:733 - 42; 10.1007/s00425-004-1316-4; PMID: 15232695
- Hwang I, Sheen J. Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature 2001; 413:383 - 9; 10.1038/35096500; PMID: 11574878
- Mira-Rodado V, Sweere U, Grefen C, Kunkel T, Fejes E, Nagy F, et al. Functional cross-talk between two-component and phytochrome B signal transduction in Arabidopsis. J Exp Bot 2007; 58:2595 - 607; 10.1093/jxb/erm087; PMID: 17545225
- Sweere U, Eichenberg K, Lohrmann J, Mira-Rodado V, Bäurle I, Kudla J, et al. Interaction of the response regulator ARR4 with phytochrome B in modulating red light signaling. Science 2001; 294:1108 - 11; 10.1126/science.1065022; PMID: 11691995
- Ishida K, Yamashino T, Mizuno T. Expression of the cytokinin-induced type-A response regulator gene ARR9 is regulated by the circadian clock in Arabidopsis thaliana. Biosci Biotechnol Biochem 2008; 72:3025 - 9; 10.1271/bbb.80402; PMID: 18997423
- Salomé PA, To JP, Kieber JJ, McClung CR. Arabidopsis response regulators ARR3 and ARR4 play cytokinin-independent roles in the control of circadian period. Plant Cell 2006; 18:55 - 69; 10.1105/tpc.105.037994; PMID: 16326927
- Ren B, Liang Y, Deng Y, Chen Q, Zhang J, Yang X, et al. Genome-wide comparative analysis of type-A Arabidopsis response regulator genes by overexpression studies reveals their diverse roles and regulatory mechanisms in cytokinin signaling. Cell Res 2009; 19:1178 - 90; 10.1038/cr.2009.88; PMID: 19621034
- Jeon J, Kim NY, Kim S, Kang NY, Novák O, Ku SJ, et al. A subset of cytokinin two-component signaling system plays a role in cold temperature stress response in Arabidopsis. J Biol Chem 2010; 285:23371 - 86; 10.1074/jbc.M109.096644; PMID: 20463025
- D’Agostino IB, Deruère J, Kieber JJ. Characterization of the response of the Arabidopsis response regulator gene family to cytokinin. Plant Physiol 2000; 124:1706 - 17; 10.1104/pp.124.4.1706; PMID: 11115887
- Müller B. Generic signal-specific responses: cytokinin and context-dependent cellular responses. J Exp Bot 2011; 62:3273 - 88; 10.1093/jxb/erq420; PMID: 21212299
- Desikan R, Hancock JT, Bright J, Harrison J, Weir I, Hooley R, et al. A role for ETR1 in hydrogen peroxide signaling in stomatal guard cells. Plant Physiol 2005; 137:831 - 4; 10.1104/pp.104.056994; PMID: 15761208
- Schütze K, Harter K, Chaban C. Bimolecular fluorescence complementation (BiFC) to study protein-protein interactions in living plant cells. Methods Mol Biol 2009; 479:189 - 202; 10.1007/978-1-59745-289-2_12; PMID: 19083187
- Walter M, Chaban C, Schütze K, Batistic O, Weckermann K, Näke C, et al. Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J 2004; 40:428 - 38; 10.1111/j.1365-313X.2004.02219.x; PMID: 15469500
- Hutchison CE, Li J, Argueso C, Gonzalez M, Lee E, Lewis MW, et al. The Arabidopsis histidine phosphotransfer proteins are redundant positive regulators of cytokinin signaling. Plant Cell 2006; 18:3073 - 87; 10.1105/tpc.106.045674; PMID: 17122069
- To JP, Kieber JJ. Cytokinin signaling: two-components and more. Trends Plant Sci 2008; 13:85 - 92; 10.1016/j.tplants.2007.11.005; PMID: 18262459
- Dortay H, Mehnert N, Bürkle L, Schmülling T, Heyl A. Analysis of protein interactions within the cytokinin-signaling pathway of Arabidopsis thaliana. FEBS J 2006; 273:4631 - 44; 10.1111/j.1742-4658.2006.05467.x; PMID: 16965536
- Horák J, Grefen C, Berendzen KW, Hahn A, Stierhof YD, Stadelhofer B, et al. The Arabidopsis thaliana response regulator ARR22 is a putative AHP phospho-histidine phosphatase expressed in the chalaza of developing seeds. BMC Plant Biol 2008; 8:77 - 95; 10.1186/1471-2229-8-77; PMID: 18625081
- Leonhardt N, Kwak JM, Robert N, Waner D, Leonhardt G, Schroeder JI. Microarray expression analyses of Arabidopsis guard cells and isolation of a recessive abscisic acid hypersensitive protein phosphatase 2C mutant. Plant Cell 2004; 16:596 - 615; 10.1105/tpc.019000; PMID: 14973164
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One 2007; 2:e718; 10.1371/journal.pone.0000718; PMID: 17684564
- To JP, Haberer G, Ferreira FJ, Deruère J, Mason MG, Schaller GE, et al. Type-A Arabidopsis response regulators are partially redundant negative regulators of cytokinin signaling. Plant Cell 2004; 16:658 - 71; 10.1105/tpc.018978; PMID: 14973166
- Leibfried A, To JP, Busch W, Stehling S, Kehle A, Demar M, et al. WUSCHEL controls meristem function by direct regulation of cytokinin-inducible response regulators. Nature 2005; 438:1172 - 5; 10.1038/nature04270; PMID: 16372013
- Dortay H, Gruhn N, Pfeifer A, Schwerdtner M, Schmülling T, Heyl A. Toward an interaction map of the two-component signaling pathway of Arabidopsis thaliana. J Proteome Res 2008; 7:3649 - 60; 10.1021/pr0703831; PMID: 18642946
- Urao T, Miyata S, Yamaguchi-Shinozaki K, Shinozaki K. Possible His to Asp phosphorelay signaling in an Arabidopsis two-component system. FEBS Lett 2000; 478:227 - 32; 10.1016/S0014-5793(00)01860-3; PMID: 10930573
- Grefen C, Donald N, Hashimoto K, Kudla J, Schumacher K, Blatt MR. A ubiquitin-10 promoter-based vector set for fluorescent protein tagging facilitates temporal stability and native protein distribution in transient and stable expression studies. Plant J 2010; 64:355 - 65; 10.1111/j.1365-313X.2010.04322.x; PMID: 20735773
- Iwama A, Yamashino T, Tanaka Y, Sakakibara H, Kakimoto T, Sato S, et al. AHK5 histidine kinase regulates root elongation through an ETR1-dependent abscisic acid and ethylene signaling pathway in Arabidopsis thaliana.. Plant Cell Physiol 2007; 48:375 - 80; 10.1093/pcp/pcl065; PMID: 17202180
- Tran LS, Urao T, Qin F, Maruyama K, Kakimoto T, Shinozaki K, et al. Functional analysis of AHK1/ATHK1 and cytokinin receptor histidine kinases in response to abscisic acid, drought, and salt stress in Arabidopsis. Proc Natl Acad Sci U S A 2007; 104:20623 - 8; 10.1073/pnas.0706547105; PMID: 18077346
- Hwang I, Chen HC, Sheen J. Two-component signal transduction pathways in Arabidopsis. Plant Physiol 2002; 129:500 - 15; 10.1104/pp.005504; PMID: 12068096
- Hass C, Lohrmann J, Albrecht V, Sweere U, Hummel F, Yoo SD, et al. The response regulator 2 mediates ethylene signalling and hormone signal integration in Arabidopsis. EMBO J 2004; 23:3290 - 302; 10.1038/sj.emboj.7600337; PMID: 15282545
- Scharein B, Voet-van-Vormizeele J, Harter K, Groth G. Ethylene signaling: identification of a putative ETR1-AHP1 phosphorelay complex by fluorescence spectroscopy. Anal Biochem 2008; 377:72 - 6; 10.1016/j.ab.2008.03.015; PMID: 18384742
- Qu X, Schaller GE. Requirement of the histidine kinase domain for signal transduction by the ethylene receptor ETR1. Plant Physiol 2004; 136:2961 - 70; 10.1104/pp.104.047126; PMID: 15466228
- Wanke D, Hohenstatt ML, Dynowski M, Bloss U, Hecker A, Elgass K, et al. Alanine zipper-like coiled-coil domains are necessary for homotypic dimerization of plant GAGA-factors in the nucleus and nucleolus. PLoS One 2011; 6:e16070; 10.1371/journal.pone.0016070; PMID: 21347358
- Grefen C, Obrdlik P, Harter K. The determination of protein-protein interactions by the mating-based split-ubiquitin system (mbSUS). Methods Mol Biol 2009; 479:217 - 33; 10.1007/978-1-59745-289-2_14; PMID: 19083185