Abstract
Gene silencing technology, such as RNA interference (RNAi), is commonly used to reduce gene expression in plant cells, and exogenous double-stranded RNA (dsRNA) can induce gene silencing in higher plants. Previously, we showed that the delivery of double-stranded DNA (dsDNA) fragments, such as PCR products of an endogenous gene sequence, into fern (Adiantum capillus-veneris) gametophytic cells induces a sequence-specific gene silencing that we termed DNAi. In this study, we used a neochrome 1 gene (NEO1) that mediates both red light-induced chloroplast movement and phototropism as a model of DNAi and confirmed that the NEO1 function was suppressed by the repression of the NEO1 gene. Interestingly, the gene silencing effect by DNAi was found in the progeny. Cytosine methylation was detected in the NEO1-silenced lines. The DNA modifications was present in the transcriptional region of NEO1, but no differences between wild type and the silenced lines were found in the downstream region of NEO1. Our data suggest that the DNAi gene silencing effect that was inherited throughout the next generation is regulated by epigenetic modification. Furthermore, the histone deacetylase inhibitor, trichostatin A (TSA), recovered the expression and function of NEO1 in the silenced lines, suggesting that histone deacetylation is essential for the direct suppression of target genes by DNAi.
Introduction
RNAi-induced sequence-specific gene silencing is an essential mechanism in plants. This process is involved in various physiological phenomena, such as the defense against viral infection, silencing of transgenes and/or endogenous genes, RNA-directed DNA methylation (RdDM), and plant development.Citation1 The RNAi mechanism has been studied using the higher plant, Arabidopsis thaliana, but RNAi has also been reported in lower plants. RNAi by both transient expression and stable transformation is a powerful tool for the functional analyses of plant genes in mosses.Citation2 In ferns, RNAi in germinating spores provides a simple single-cell system to study gene function.Citation3 Transcriptional gene silencing and de novo DNA methylation of a transgene-homologous promoter occur if the dsRNA contains promoter sequences. Indeed, an RNAi component is involved in the de novo DNA methylation at cytosine residues in a phenomenon known as RNA-directed DNA methylation.Citation4 DNA methylation is found at cytosines in three different sequence contexts, CG, CNG (where N is any base), and asymmetric CHH (where H = A, T, or C). DNA methylation is maintained by three DNA methyltransferases. Cytosine DNA methyltransferase 1 (MET1) encodes a DNA methyltransferase that is orthologous to a mouse Dnmt1.Citation5 The maintenance activity for DNA methylation of MET1 replicates CG DNA methylation, even when the initial trigger for the DNA methylation is genetically removed.Citation6,Citation7 However, non-CG methylation is mostly maintained by the other two methyltransferases, CMT3 and DRM2. Non-CG DNA methylation is inherited in different ways and requires a continuously active signal to target the regions of DNA methylation.Citation8 In the case of CNG and CHH methylation, the signal seems to come from histones that are associated with the DNA region. Non-CG methylation also often requires histone H3K9 dimethylation in Arabidopsis.Citation9,Citation10 Promoters silenced by RdDM are maintained by histone RPD3-type histone deacetylase AtHDA6,Citation11 which is a member of a large gene family of histone deacetylases (HDACs) in the Arabidopsis genome. AtHDA6 knockout plants exhibit the transcriptional reactivation of promoters targeted by RdDM, despite the continuous presence of the silencing-inducing RNA signal.Citation11
In our previous study, we reported that the delivery of dsDNA fragments, which were homologous to an endogenous gene, by particle bombardment efficiently caused a knockout phenotype of the corresponding gene in Adiantum capillus-veneris. We termed this phenomenon DNA interference (DNAi), based on the analogy to RNAi. PCR-amplified gene fragments were sufficient to trigger DNAi, and the simultaneous delivery of several different gene fragments effectively silenced all of the gene functions. DNAi-induced gene silencing was shown to spread throughout the entire plant (systemic silencing) from a bombarded cell, where the target gene was silenced.Citation12 In this report, we determined that the DNAi inheritability, at least in part, was dependent on epigenetic regulation, including DNA methylation.
Results
The gene silencing effect by DNAi is passed on to the next generation
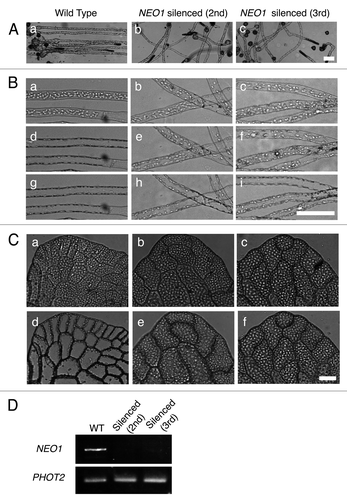
Our previous study showed that the introduction of DNA fragments of a NEO1 intronless gene, encoding a photoreceptor mediating red light-induced phototropism and chloroplast relocation, gave rise to the same phenotype as NEO1 deficient mutant plants.Citation13 We investigated whether the gene silencing effect by DNAi remained in the next generation by microscopic observation and the RT-PCR analysis of NEO1 expression. Spores obtained from the sporophytes that developed on the prothalli bombarded with partial gene fragments of NEO1 (see the schematic diagram in ) (i.e., the spores are the 2nd generation of a line silenced by DNAi) were cultured under unilateral red light to analyze the red light-induced phototropism. The direction of growth of the protonemata that germinated from the spores was variable, regardless of the direction of red light, whereas the protonemata from wild type spores grew straight, toward the red light source. Interestingly, the 3rd generation protonemata grown from the spores obtained from the 2nd generation sporophytes also showed a phenotype similar to the 2nd generation (). The chloroplast relocation movement was also examined in the protonemata and prothalli of the 2nd and the 3rd generations of the silenced lines. The protonemata were irradiated horizontally with a vertically vibrating polarized red or blue light. In the wild type protonemata, the chloroplasts accumulated on both sides of the cell. However, in the protonemata of the 2nd and the 3rd generations, the chloroplasts distributed randomly under the polarized red light (). A similar deficiency of chloroplast relocation response under polarized red light was observed in the prothallial cells of the 2nd and 3rd generations (). According to an RT-PCR analysis, it was reported that the DNAi effect was caused by the downregulation of the transcriptional level of a target gene.Citation12 To confirm the red light results, we investigated the level of NEO1 transcript by RT-PCR analysis in the gametophytes of each generation. The blue light photoreceptor, phototropin2 (PHOT2), was used as an internal control. All of the gametophytes had almost the same expression level of PHOT2 in the 2nd and 3rd generations of the silenced lines, as well as in the wild type gametophytes. However, the NEO1 transcripts were barely detectable in these descendants of the silenced lines, although strongly detected in the wild type (). These results indicate that the effect of DNAi on NEO1 expression remained over generations.
Figure 2. The cytosine methylation profiles in silenced lines. Top: the NEO1 gene structure. The thick, thin solid and broken lines represent the ORF, UTR and promoter regions of the NEO1 gene, respectively. The dotted line and double arrows represent the PCR fragment for the DNAi and the regions for the Chop-PCR amplification, respectively. The small arrow indicates the poly-adenylation site. Bottom: Chop-PCR analyses. After genomic DNA was digested with a methylation-sensitive restriction enzyme, HpaII or AluI, or with a methylated DNA-specific restriction enzyme, McrBC, it was used for the amplification of each NEO1 region (0, I, II, III, and IV) corresponding to a region of NEO1, as indicated in the upper panel. The amplification of non-digested genomic DNA was used for a control.
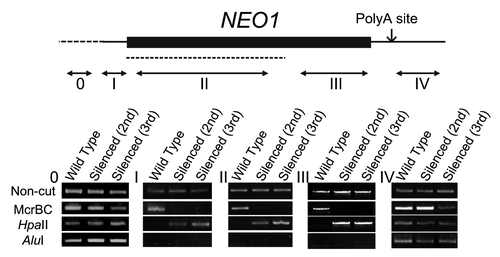
Figure 1. The inheritance of the DNAi-induced NEO1 phenotype over generations. (A) Red light-induced phototropism in the protonemata of wild type (a), the 2nd generation (b), and the 3rd generation (c) of a NEO1 silenced line. The protonemata were cultured for 10 d under unilateral red light (10.7 µmol m−2 s−1) from the right-hand side. Scale bar: 100 µm. (B) Chloroplast relocation movement in the protonemata of wild type (a, d, and g), the 2nd (b, e, and h) and 3rd (c, f, and i) generations of NEO1 silenced lines. Protonemata, precultured under a weak white light (20~30 µmol m−2 s−1, a, b, and c), were irradiated horizontally for 5 h with a vertically vibrating red (d, e, and f) or blue (g, h, and i) polarized light (red = 10.7 µmol m−2 s−1, blue = 6.2 µmol m−2 s−1). Bar: 100 µm. (C) Chloroplast relocation movements in the prothalli of wild type (a and d), the 2nd (b and e) and 3rd (c and f) generations of the NEO1 silenced lines. The chloroplast movement was induced by irradiation with a low fluence-rate white light (20~30 µmol m−2 s−1, a, b, and c) from above and then a vertically vibrating polarized red light (10.7 µmol m−2 s−1, d, e, and f) horizontally for 5 h. Bar: 50 μm. (D) The gene expression analysis of NEO1 in NEO1-silenced lines by RT-PCR. Total RNA was prepared from the prothalli of wild type and NEO1-silenced lines of the 2nd and 3rd generations. PHOT2 was used as an amplification control.
Genomic cytosine methylation exists in the transcriptional region of NEO1 gene in DNAi-induced fern
RNA-directed DNA methylation (RdDM) is found in the gene silencing induced by 24-nt small RNAs.Citation14 Therefore, cytosine methylation may be detected in lines silenced by DNAi. We attempted to find cytosine methylation in the silenced lines by a Chop-PCR method using a methylated DNA specific enzyme, McrBC, and the methylation-sensitive enzymes, HpaII and AluI. In these methods, non- or hypo-methylated DNA templates survive under McrBC treatment and are amplified by the ensuing PCR, but hypermethylated DNA is “chopped” by McrBC, so that no PCR product is amplified. In contrast, non-methylated DNA is chopped by HpaII, so that no PCR products are obtained. When genomic DNA of the 2nd generation silenced line was treated with McrBC, no DNA was amplified by PCR (), whereas HpaII-treated genomic DNA was amplified, suggesting that cytosine methylation occurred in the genomic NEO1 region of the silenced lines. To delineate the NEO1 genomic regions where cytosine was methylated, the 5′UTR, the ORF region, including or excluding the DNA sequence used for the DNAi, the 3′ downstream region of NEO1, and four regions of NEO1 were analyzed. The Chop-PCR results showed that cytosine methylation was present in the transcriptional region of the silenced lines (, regions I, II, and III). A PCR fragment was amplified from the 3′ downstream region of the wild type and silenced lines following either McrBC or HpaII treatment, indicating that this region contained both methylated and non-methylated cytosines (, region IV). These results suggest that the gene silencing by DNAi was mediated by the cytosine methylation of the transcriptional region in the silenced lines. Interestingly, although the methylation of promoter region is critical for gene silencing in other plants or animals, DNA methylation in the promoter region of NEO1 gene was not detected, suggesting that the DNA-induced DNA methylation might be somewhat different from RdDM (, region 0). Furthermore, the HpaII and AluI recognition sites contain CG and CNG, and CNG and CHH, respectively. Our Chop-PCR analysis showed that there was no amplicon when the NEO1 transcriptional regions were treated with AluI, although a PCR fragment was amplified after HpaII treatment in the silenced lines, suggesting that only CG sites were methylated, but non-CG sites were not methylated, in the NEO1 gene of the NEO1 silenced lines ().
Histone deacetylase inhibitor rescues the NEO1 function in the silenced line, but cytosine methylation reduction does not
In yeast, histone subunit 3 is deacetylated at the 9th lysine by a histone deacetylase (HDAC),Citation15 and trichostatin A (TSA) is an HDAC inhibitor. In Arabidopsis, cytosines are methylated by DNA methyltransferases, such as MET1, DRM1, 2 and CMT3, and DNA methylation is inhibited by 5-azacytidine (AZA), which is a cytosine analog.Citation14 In this study, we analyzed whether TSA rescued the NEO1 function in DNAi silenced lines, based on the chloroplast photorelocation movement in the gametophytes by red light irradiation. When treated with TSA, the chloroplasts moved to the anticlinal wall under polarized red light in the silenced lines, although they were at the cell surface before treatment, suggesting that TSA has a positive function in prothalli (). This result suggests that TSA treatment cancelled the dimethylation of H3K9 in the silenced lines and that histone deacetylation may recruit histone dimethylation in the DNAi of Adiantum. Because the NEO1 expression was suppressed in both of the silenced lines (), if TSA could rescue the NEO1 function, the NEO1 transcriptional level should be recovered in the silenced line upon TSA treatment. When total RNA, extracted from the silenced lines, was treated with TSA, the NEO1 transcript was strongly detected by RT-PCR analysis (). Cytosine methylation was detected and the histone dimethylation was predicted in the silenced lines but not in wild type (Figs. Two and 3). It is possible that the histone dimethylation had a synergistic influence on cytosine methylation. However, Chop-PCR showed that cytosine methylation by DNAi was not cancelled by the TSA treatment (). Conclusively, histone modification is independent from DNA methylation in the DNAi system.
Figure 3. The effects of trichostatin A (TSA) and 5-azacytidine (AZA) on NEO1-silenced lines. (A) Chloroplast relocation movement in silenced lines treated with or without TSA or AZA. Gametophytes were treated with 5 µg ml−1 TSA for 2–3 weeks (a, c, e, g, I, and k) or 500 µM AZA (b, d, f, h, j, and l) for 2–3 weeks. Chloroplast movement was induced by low fluence-rate white light, directed from above (20~30 µmol m−2 s−1, a, b, c, d, e, and f), and then vertically vibrating polarized red light (10.7 µmol m−2 s−1, g, h, i, j, k, and l) from a horizontal direction for 5 h. Bar = 50 μm. (B) RT-PCR analysis in DNAi lines with or without TSA or AZA. Total RNAs were prepared from prothalli treated with 5 µg ml−1 TSA or 500 µM AZA for 2–3 weeks. PHOT2 was used as an amplification control. (C) Chop analysis with or without TSA or AZA. Genomic DNA obtained from prothalli that were incubated with 5 µg ml−1 TSA or 500 µM AZA for 2–3 weeks were used as the template for Chop-PCR.
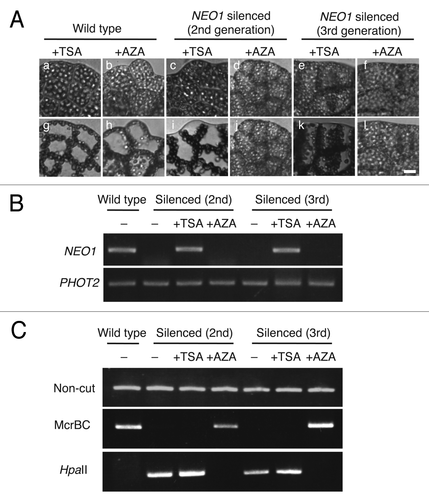
To confirm whether 5-azacytidine (AZA) could rescue the NEO1 function in the DNAi lines, the chloroplast relocation movement was analyzed by vertically vibrating polarized red light in the gametophytes of the silenced lines under treatment with AZA. However, the chloroplast did not change their location and remained at the cell surface (). The transcriptional levels of NEO1 and the cytosine methylation in the silenced lines were also studied with or without AZA. NEO1 expression level did not change regardless of AZA existence, meaning that the histon modification was maintained (). In contrast, cytosine methylation was cancelled by AZA treatment (). Taken together, both DNA methylation and histone modification are obviously important for DNAi induction. In particular, histone modification is essential for the direct suppression of a target gene by DNAi, although the function of DNA methylation is still unknown.
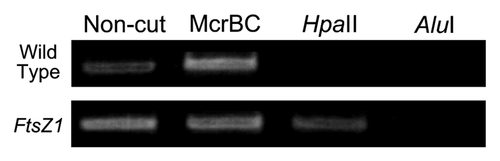
DNAi was effective in all genes so far tested in A. capillus-veneris, but the DNAi effective period is different depending on genes. Further, in all genes tested, except NEO1 gene, DNAi effects were non-inheritable, so that wild type phenotype in each gene was recovered within their gametophyte generation. Thus we tested whether DNA methylation occurred in DNAi-induced but non-inheritable gene using FtsZ1Citation12 gene that is involved in chloroplast division. Many gametophytes with large chloroplasts were obtained in FtsZ1 bombarded gametophytes, although the silenced levels of this gene are different in each gametophyte even in different cells of one gametophyte, so that the size of chloroplasts were various among cells. DNA methylation was detected using almost all gametophytes screened as one sample for Chop-PCR analysis (). Both methylated and non-methylated FtsZ1 genes were present in this population, suggesting that both DNAi–induced and -non-induced cells co-existed in the population. As a result, DNAi-induced FtsZ1 gene silencing is depending on DNA methylation.
Figure 4. The cytosine methylation profiles in FtsZ gene-silenced lines in Chop-PCR analyses. After genomic DNA was digested with a methylation-sensitive restriction enzyme, HpaII or AluI, or with a methylated DNA-specific restriction enzyme, McrBC, the digested D|NA was used for the amplification. Other details were the same as in .
Discussion
Heritability of DNAi over generations
We demonstrated that the DNAi effect was transmitted to the next generation and that the effect was associated with the expression of NEO1 (). The PCR-amplified DNA fragments of the transcriptional region commonly called the ‘gene body’ were used for the induction of gene silencing by DNAi (). The heritability of RNAi has been reported in tobacco,Citation7 in which 35S::GFP and tobacco mosaic virus (TMV) were used as a reporter and an inducer for gene silencing, respectively. When a TMV containing the gene body of GFP was infected into cells, the RNAi-triggered gene silencing was not inherited by the next generation. However, the RNAi effect induced by a TMV containing a 35S promoter was inherited. The same phenomena have been confirmed in Arabidopsis.Citation6 Therefore, RNAi heritability was believed to be limited to a promoter region of a target gene as an RNAi inducer. In contrast, DNAi by PCR derived fragments from the gene body of a target gene were inheritable in fern NOE1 gene silencing. It is noteworthy that not the promoter region but the gene body of the target gene is useful as an inducer of DNAi transmission to the next generation in A. capillus-veneris. Further, since the DNA methylation occurs in FtsZ1 gene (), inheritance of other fern genes may be possible. However, the DNAi effects of FtsZ1 and other genes tested were non-inheritable and wild type phenotype of these genes recovered within their gametophyte generation, suggesting that a mechanism of DNA de-methylation might exist in fern cells and the efficiency of DNA de-methylation might be affected by the number or the length of introns.
Histone modification in DNAi
Histone deacetylation was predicted in NEO1-silenced lines by inhibitor experiments. This deacetylation was correlated with the gene expression of NEO1 ( and ). The HDAC inhibitor, TSA, cancelled the DNAi effect and recovered NEO1 expression (). DNA methylation was detected in the 5′ UTR and coding regions of NEO1, but not in the 3′ downstream region, suggesting that the DNAi effect was not transmitted to neighboring genes. Stout et al.Citation3 developed an RNAi method in fern using the direct delivery of dsRNA into spores through imbibitions, and it was suggested that a gene silencing system by RNAi machinery exists in Ceratopteris richardii.Citation3 Indeed, we found genes involved in small RNA biogenesis and epigenetic regulation, such as HDAC, in an Adiantum EST database (http://togodb.dbcls.jp/acest/).Citation16 Ferns may have an RNAi-dependent epigenetic regulation system similar to higher plants, because many of the conserved genes for RNAi systems are present in fern species. Additionally, Ceratopteris richardii has a DNAi system.Citation17 Therefore, ferns have two possible systems: DNAi and RNAi. These systems may share genes involved in epigenetic regulation, because the effects of both RNAi and DNAi are regulated by HDAC.
We also showed the possibility of histone modification in the silenced lines using, HDAC inhibitor, TSA that cancelled the effect of DNAi (). Cytosine methylation by RNAi in plants is associated with histone modification to suppress gene expression.Citation11 Acetylation at the 9th Lys in histone subunit 3 (H3K9acetyl) is related to transcriptional activation,Citation18 and their dimethylation at the 9th Lys (H3K9dimethyl) is related to the suppression of gene expression, which includes heterochromatin formation.Citation19 Histone deacetylation leads to histone H3K9 dimethylation in yeast and Arabidopsis,Citation15,Citation18 and cytosine methylation is associated with H3K9 dimethylation in Arabidopsis.Citation20 If the histone modification is associated with DNA methylation in DNAi, histones may be deacetylated and/or dimethylated in the silenced lines. So, we tried repeatedly chromatin immunoprecipitation experiments to know the status of the histones, but it was not successful because the enough amount of purified DNA could not be obtained from the fern prothalli. Fern gametophytes are difficult material for biochemistry because (1) the cells are large and mostly vacuolated, (2) collecting enough amount of tissues are really hard work, (3) further worse polysaccharides are an obstacle for DNA purification.
DNA methylation in NEO1-silenced lines
In plants, the transcriptional gene silencing that results from RNAi in promoter regions is correlated with an increase in promoter methylation.Citation21 DNA methylation is the hallmark of RNA mediated silencing in plants. Here, we showed that cytosine methylation was found in the gene body of a NEO1-silenced line but not in wild type gametophytes (). In general, DNA methylation is inherited by the next generation. Because the DNA methylation at a promoter region is inheritable in RNAi-dependent gene silencing,Citation11 it is plausible that DNAi may use DNA methylation as an inheritable memory (epigenetic memory), similar to RNAi-silencing mechanisms. However, the de-methylation of NEO1-silenced lines by AZA could not cancel the gene silencing in Adiantum, although the DNA de-methylation by AZA cancelled the gene silencing by RNAi ().Citation22-Citation24 These studies suggest that DNA methylation and histone deacetylation are synergistic in the gene silencing of RNAi in Arabidopsis. However, our results showed that histone modification is independent of DNA methylation in Adiantum (). The differences between DNAi and RNAi may or may not be explained by the developmental stage of the plants or the timing of DNA methylation in the silenced lines. When AZA was employed for RNAi experiments in higher plants, such as Arabidopsis and maize, sporophytic diplophase plants were used.Citation14,Citation25 In contrast, we used fern haplophase gametophytes in our DNA methylation analysis for DNAi. It is also plausible that DNA methylation might be used to trigger DNAi processes.
Genome-wide analyses of DNA methylation in Arabidopsis have shown that most genes have unmethylated promoters, yet 20–30% of the genes show DNA methylation in the gene body.Citation26 However, the role of DNA methylation in the gene body is still elusive. Gene body methylation in DNAi-dependent NEO1-silenced lines might provide a clue to resolve this function.
The role of DNAi in fern
The RNAi system has an important role in plants. Gene silencing by RNAi is an inducible immunity system that specifically targets viral infection: viral infection triggers an RNAi system in plants to destroy the virus RNAs.Citation27,Citation28 In addition, the integrated DNA from a virus and/or transposon is inactivated by DNA methylation through a plant RNAi system.Citation29,Citation30 What, then, is the function of DNAi in ferns? It is believed that ferns may have pathogenic resistance systems against fungi and viruses, although we could not find any report concerning the infection of a DNA virus in ferns. However, the DNAi system may still be useful in the protection against a DNA virus infection. The combination of DNAi and RNAi systems in ferns might play an important role for supporting a high pathogenic resistance.
Materials and Methods
Plant materials
Aseptically cultivated prothalli of Adiantum capillus-veneris L. were cut into pieces with a Polytron (PT1200, Kinematica), and the fragments were grown under fluorescent lamps (FL40SD Toshiba Lighting and Technology Corp.) (20–30 µmol m−2 s−1) for 3 weeks and used for DNA and histone analyses.Citation31 For particle bombardment, the regenerated prothalli were placed on the surface of an agar plate containing White’s medium solidified with 0.5% agar (Funakoshi) in a 6 cm plastic Petri dish and aseptically bombarded.Citation32
Preparation of silenced lines by DNAi
The DNAi procedure was performed according to Kawai-Toyooka et al., 2004. Prothalli were subjected to osmotic pretreatment by placing them on the surface of a medium solidified with 0.5% agar that contained 0.25 M mannitol for 1 h prior to the bombardment, performed with a Biolistic PDS-1000/He Particle Delivery System (Bio-Rad). The DNA delivery conditions were as described previously.Citation12 The PCR amplification of the NEO1 and FtsZ1 gene for bombardment was performed with ExTaq PCR kit (Takara). The amount of DNA delivered was 0.48 pmol for each bombardment. After bombardment, the prothalli were incubated in darkness for 1 d and then cultured under a white fluorescent lamp, as described above, for 4 d. The prothalli were then transferred to a selective medium (White’s medium containing 10 µg ml–1 hygromycin B) and cultured under white light for 9–14 d. After that, they were transferred to White’s medium without hygromycin B, and the phenotypes of phototropism and chloroplast relocation movement were observed. Prothalli defective in the above phenomena were selected and cultured under white light as the first generation (1st) of silenced lines. These prothalli were cultivated until sporophytes (diploid) emerged after self-fertilization. Spores collected from these sporophytes and the gametophytes grown from these spores were considered the second generation (2nd) of the silenced lines. The spores and gametophytes obtained from the sporophytes of the 2nd generation were considered the third generation (3rd). To maintain gametophyte lines of each generation, the fragmentation and subculture of the gametophytes were repeated every 3 weeks until use. All of the procedures were performed at 25°C.
Phenotypic analysis
To obtain protonemal cells for phototropic analysis, spores were sterilized with a 0.5% (volume/volume) bleach solution (Wako Pure Chemical Industries) for 30 sec and sown on the surface of medium containing White’s medium solution solidified with 0.5% agar (Funakoshi) in a 3 cm glass Petri dish and covered with a coverslip (3 mm in width). The spores were cultured under unilateral red light, which was obtained from the fluorescence lamp described above and a red plastic filter (Shinkolite A no. 102, Mitsubishi Rayon), for approximately 10.7 µmol m−2 s−1 for 7 d. To observe chloroplast movement in protonemata, the protonemata were incubated for 5 h under polarized unilateral red (~10.7 µmol m−2 s−1) or blue light (~6.2 µmol m−2 s−1). The polarized light was obtained by passing light through a linear polarizer (HN38, Polaroid Corp. of Japan). To observe the chloroplast movement in prohtallial cells, prothalli were cultivated under white light for 3 weeks after fragmentation and then transferred to a Petri dish (3 cm in diameter) and covered with a coverslip, as described above. The light conditions for the chloroplast movement induction in the prothalli were the same as those for the protonemata.
Inhibitor treatments
After being cut into pieces, prothalli were grown under white light for 1 week and then transferred to a new medium containing 5 µg ml−1 trichostatin A (TSA) or 500 µM 5-azacytidine (AZA). After 2–3 weeks of cultivation under white light, the prothalli were used for RT-PCR analysis or transferred to a glass Petri dish (3 cm in diameter) and covered with a coverslip (3 mm in width) for chloroplast observation. Images of the prothalli were taken with a digital camera (ORCA-ER, Hamamatsu Photonics) equipped on a microscope (Axio imager Z1, Carl Zeiss).
Genomic DNA and total RNA preparation
Genomic DNA and total RNA, derived from the gametophytes, were isolated by a cetyltrimethylammonium bromide (CTAB) method.Citation33 After the gametophytes were ground with a mortar and pestle under liquid nitrogen condition, the powder was incubated with 10 times the volume of a 2 x CTAB solution (2% [weight/volume] CTAB, 0.1 M TRIS-HCl, pH 8.0, 20 mM EDTA, and 1.4 M NaCl) containing 5% (volume/volume) 2-mercaptoethanol at 60°C. Chloroform extractions were repeated three times. The DNA/RNA mixtures were then precipitated with isopropanol, and the precipitates were dissolved in TE. Total RNA was precipitated twice with 1/4 volume of 10 M LiCl. The genomic DNA was precipitated with 3 volumes of ethanol, using the LiCl-soluble fraction, and was purified by the DNAeasy kit (Qiagen), following the manufacturer’s instructions. The total RNA or genomic DNA precipitates were dissolved in diethylpyrocarbonate (DEPC)–treated water or sterilized water (nuclease free), respectively.
RT-PCR analysis
The total RNA samples were treated with DNase (Takara) and then reverse-transcribed using an oligo(dT) primer and the SUPER-SCRIPTIII preamplification system for the first-strand cDNA synthesis (Invitrogen). The cDNA reaction mixture was used as the template for PCR. The PCR was performed with the MightyAmp (Takara). PCR conditions were a preincubation for 2 min at 98°C, followed by 40 cycles of denaturation for 15 sec at 98°C, annealing for 30 sec at 65°C, and extension for 2 min at 68°C. PHOT2 was used as the internal control.
DNA methylation assay
The DNA methylation status was analyzed by Chop-PCR. Genomic DNA (200 ng) was digested with a methylation-sensitive restriction enzyme HpaII and AluI or a methylated DNA–digesting enzyme, McrBC, overnight. The digested DNA was used for the amplification of NEO1 using the MightyAmp (Takara) and the indicated primers. PCR was performed for 2 min at 98°C, followed by 40 cycles of 10 sec at 98°C, 15 sec at 65°C, and 1.5 min at 28°C. The PCR products were then subjected to electrophoresis.
| Abbreviations: | ||
| DNAi | = | DNA interference |
| RNAi | = | RNA interference |
| dsRNA | = | double-strand RNA |
| dsDNA | = | double-strand DNA |
| TSA | = | trichostatin A |
| AZA | = | 5-azacytidine |
| HDAC | = | histone deacetylase |
| RdDM | = | RNA-directed DNA methylation |
| H3K9 | = | histone subunit 3 at 9th Lys |
| H3K14 | = | histone subunit 3 at 14th Lys |
| TMV | = | tobacco mosaic virus |
| CTAB | = | cetyltrimethylammonium bromide |
| DEPC | = | diethylpyrocarbonate |
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was supported in part by the Japan Society for the Promotion of Science (JSPS 20227001 and 22657016 to M.W.) and a Research Fellowship for Young Scientists (to H.T.).
Acknowledgments
We express our sincere thanks to Dr. Noriyuki Suetsugu for helpful discussions.
References
- Baulcombe D. RNA silencing in plants. Nature 2004; 431:356 - 63; 10.1038/nature02874; PMID: 15372043
- Bezanilla M, Pan A, Quatrano RS. RNA interference in the moss Physcomitrella patens.. Plant Physiol 2003; 133:470 - 4; 10.1104/pp.103.024901; PMID: 14555775
- Stout SC, Clark GB, Archer-Evans S, Roux SJ. Rapid and efficient suppression of gene expression in a single-cell model system, Ceratopteris richardii.. Plant Physiol 2003; 131:1165 - 8; 10.1104/pp.016949; PMID: 12644667
- Matzke MA, Birchler JA. RNAi-mediated pathways in the nucleus. Nat Rev Genet 2005; 6:24 - 35; 10.1038/nrg1500; PMID: 15630419
- Finnegan EJ, Dennis ES. Isolation and identification by sequence homology of a putative cytosine methyltransferase from Arabidopsis thaliana.. Nucleic Acids Res 1993; 21:2383 - 8; 10.1093/nar/21.10.2383; PMID: 8389441
- Aufsatz W, Mette MF, van der Winden J, Matzke AJM, Matzke M. RNA-directed DNA methylation in Arabidopsis. Proc Natl Acad Sci U S A 2002; 99:Suppl 4 16499 - 506; 10.1073/pnas.162371499; PMID: 12169664
- Jones L, Ratcliff F, Baulcombe DC. RNA-directed transcriptional gene silencing in plants can be inherited independently of the RNA trigger and requires Met1 for maintenance. Curr Biol 2001; 11:747 - 57; 10.1016/S0960-9822(01)00226-3; PMID: 11378384
- Chan SWL, Henderson IR, Jacobsen SE. Gardening the genome: DNA methylation in Arabidopsis thaliana.. Nat Rev Genet 2005; 6:351 - 60; 10.1038/nrg1601; PMID: 15861207
- Bartee L, Malagnac F, Bender J. Arabidopsis cmt3 chromomethylase mutations block non-CG methylation and silencing of an endogenous gene. Genes Dev 2001; 15:1753 - 8; 10.1101/gad.905701; PMID: 11459824
- Jackson JP, Lindroth AM, Cao X, Jacobsen SE. Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature 2002; 416:556 - 60; 10.1038/nature731; PMID: 11898023
- Aufsatz W, Mette MF, van der Winden J, Matzke M, Matzke AJM. HDA6, a putative histone deacetylase needed to enhance DNA methylation induced by double-stranded RNA. EMBO J 2002; 21:6832 - 41; 10.1093/emboj/cdf663; PMID: 12486004
- Kawai H, Kanegae T, Christensen S, Kiyosue T, Sato Y, Imaizumi T, et al. Responses of ferns to red light are mediated by an unconventional photoreceptor. Nature 2003; 421:287 - 90; 10.1038/nature01310; PMID: 12529647
- Kawai-Toyooka H, Kuramoto C, Orui K, Motoyama K, Kikuchi K, Kanegae T, et al. DNA interference: a simple and efficient gene-silencing system for high-throughput functional analysis in the fern adiantum.. Plant Cell Physiol 2004; 45:1648 - 57; 10.1093/pcp/pch186; PMID: 15574841
- Chang S, Pikaard CS. Transcript profiling in Arabidopsis reveals complex responses to global inhibition of DNA methylation and histone deacetylation. J Biol Chem 2005; 280:796 - 804; PMID: 15516340
- Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 2001; 292:110 - 3; 10.1126/science.1060118; PMID: 11283354
- Yamauchi D, Sutoh K, Kanegae H, Horiguchi T, Matsuoka K, Fukuda H, et al. Analysis of expressed sequence tags in prothallia of Adiantum capillus-veneris.. J Plant Res 2005; 118:223 - 7; 10.1007/s10265-005-0209-3; PMID: 15940394
- Rutherford G, Tanurdzic M, Hasebe M, Banks JA. A systemic gene silencing method suitable for high throughput, reverse genetic analyses of gene function in fern gametophytes. BMC Plant Biol 2004; 4:6; 10.1186/1471-2229-4-6; PMID: 15090074
- Chen ZJ, Tian L. Roles of dynamic and reversible histone acetylation in plant development and polyploidy. Biochim Biophys Acta 2007; 1769: 295-307.
- Lippman Z, Gendrel AV, Black M, Vaughn MW, Dedhia N, McCombie WR, et al. Role of transposable elements in heterochromatin and epigenetic control. Nature 2004; 430:471 - 6; 10.1038/nature02651; PMID: 15269773
- Bernatavichute YV, Zhang X, Cokus S, Pellegrini M, Jacobsen SE. Genome-wide association of histone H3 lysine nine methylation with CHG DNA methylation in Arabidopsis thaliana.. PLoS One 2008; 3:e3156; 10.1371/journal.pone.0003156; PMID: 18776934
- Kooter JM, Matzke MA, Meyer P. Listening to the silent genes: transgene silencing, gene regulation and pathogen control. Trends Plant Sci 1999; 4:340 - 7; 10.1016/S1360-1385(99)01467-3; PMID: 10462766
- Chen ZJ, Pikaard CS. Epigenetic silencing of RNA polymerase I transcription: a role for DNA methylation and histone modification in nucleolar dominance. Genes Dev 1997; 11:2124 - 36; 10.1101/gad.11.16.2124; PMID: 9284051
- Kumpatla SP, Teng W, Buchholz WG, Hall TC. Epigenetic transcriptional silencing and 5-azacytidine-mediated reactivation of a complex transgene in rice. Plant Physiol 1997; 115:361 - 73; 10.1104/pp.115.2.361; PMID: 9342860
- Lawrence RJ, Earley K, Pontes O, Silva M, Chen ZJ, Neves N, et al. A concerted DNA methylation/histone methylation switch regulates rRNA gene dosage control and nucleolar dominance. Mol Cell 2004; 13:599 - 609; 10.1016/S1097-2765(04)00064-4; PMID: 14992728
- Yang F, Zhang L, Li J, Huang J, Wen R, Ma L, et al. Trichostatin A and 5-azacytidine both cause an increase in global histone H4 acetylation and a decrease in global DNA and H3K9 methylation during mitosis in maize. BMC Plant Biol 2010; 10:178; 10.1186/1471-2229-10-178; PMID: 20718950
- Zhang X, Yazaki J, Sundaresan A, Cokus S, Chan SWL, Chen H, et al. Genome-wide high-resolution mapping and functional analysis of DNA methylation in arabidopsis. Cell 2006; 126:1189 - 201; 10.1016/j.cell.2006.08.003; PMID: 16949657
- Covey SN, Al-Kaff NS, Turner ALDS. Plants combat infection by gene silencing. Nature 1997; 385:781 - 2; 10.1038/385781a0
- Ratcliff F, Harrison BD, Baulcombe DC. A similarity between viral defense and gene silencing in plants. Science 1997; 276:1558 - 60; 10.1126/science.276.5318.1558; PMID: 18610513
- Brough CL, Gardiner WE, Inamdar NM, Zhang XY, Ehrlich M, Bisaro DM. DNA methylation inhibits propagation of tomato golden mosaic virus DNA in transfected protoplasts. Plant Mol Biol 1992; 18:703 - 12; 10.1007/BF00020012; PMID: 1558945
- Tsukahara S, Kobayashi A, Kawabe A, Mathieu O, Miura A, Kakutani T. Bursts of retrotransposition reproduced in Arabidopsis. Nature 2009; 461:423 - 6; 10.1038/nature08351; PMID: 19734880
- Tsuboi H, Suetsugu N, Kawai-Toyooka H, Wada M. Phototropins and neochrome1 mediate nuclear movement in the fern Adiantum capillus-veneris.. Plant Cell Physiol 2007; 48:892 - 6; 10.1093/pcp/pcm057; PMID: 17507389
- Tsuboi H, Yamashita H, Wada M. Chloroplasts do not have a polarity for light-induced accumulation movement. J Plant Res 2009; 122:131 - 40; 10.1007/s10265-008-0199-z; PMID: 19037581
- Kanegae T, Wada M. Isolation and characterization of homologues of plant blue-light photoreceptor (cryptochrome) genes from the fern Adiantum capillus-veneris.. Mol Gen Genet 1998; 259:345 - 53; 10.1007/s004380050821; PMID: 9790588