Abstract
Small non-coding RNAs (sRNAs) emerge as exquisite molecules that are guided for transcriptional and posttranscriptional gene regulation in eukaryotes. As one class of most important sRNAs in plants, trans-acting small interfering RNAs (ta-siRNAs) initiate from microRNA (miRNA) – mediated cleavage of TAS gene transcripts and subsequently are stabilized by SUPPRESSOR OF GENE SILENCING3 (SGS3) and converted to double–stranded RNA (dsRNA) by the actions of RNA-DEPENDENT RNA POLYMERASE6 (RDR6). Generally, these dsRNAs are processed by DICER-LIKE4 (DCL4) and recruited into ARGONAUTE 1 (AGO1) complexes to posttranscriptionally regulate target genes by mRNA cleavage in trans. In a recent study, we discovered a non-canonical ta-siRNAs pathway: Starting from the miRNA-guided cleavage site, the dsRNAs are processed by DCL1 into 21-nt siRNAs, which associate with AGO4/6 complexes to direct DNA methylation in cis. Together with previous results that miRNAs can be produced by DCL3, loaded into AGO4 and trigger epigenetically regulation of target genes, these findings indicate much complex biogenesis, effector and action pathways exist in plant sRNAs kingdom.
Plant occupy small silencing RNAs, containing microRNAs (miRNAs) and small interfering RNAs (siRNAs), to exquisitely regulate target genes in a wide range of development processes and responses to abiotic and biological stresses. So far, plant endogenous siRNAs have been reported as three subcategories: heterochromatic siRNAs (hc-siRNAs), natural antisense transcript-derived siRNAs (nat-siRNAs) and trans-acting-siRNAs (ta-siRNAs).Citation1 Typically, miRNAs negatively regulate targets by mRNA cleavage and translation inhibition in trans; hc-siRNAs silence transponson and repeat elements though DNA methylation and histone modification in cis; nat-siRNAs are induced when plants grow in abnormal conditions and cause the silencing of antisense transcripts via disruption of mRNA stability in cis; the silencing fashions of ta-siRNAs like miRNAs, which usually modulate targets as slicers in trans.Citation2
DCLs and AGOs are recognized as key players in sRNA biogenesis and effector pathways. Plants harbor four classes of DCL proteins and three clades of AGO proteins. A decade ago, it is known that DCL1 is responsible for miRNAs formation from hairpin structures precursors and AGO1 is the effector complexes essential for miRNA-mediated targets mRNA cleavage, since dcl1 and ago1 mutants displayed pleiotropic phenotypes as loss of miRNA activities.Citation3,Citation4 Afterwards, people found the biogenesis of some newly evolved miRNAs in Arabidosis requires DCL4 and miRNA can function as translation repressors through AGO1, which provided insights for wide miRNA biogensis and action pathways.Citation5,Citation6 Recently, our studies in rice revealed that a class of 24-nt long miRNAs are produced by DCL3 and sorted into AGO4 clades proteins to direct DNA methylation for targets epigenetic regulation, which greatly expands the knowledge of complex miRNA production and effect pathways.Citation7
As a plant-specific siRNAs, ta-siRNAs appear to be hallmark of secondary siRNAs, which initiate from miRNA-cleavaged TAS1–4 gene transcripts.Citation8 These cleavage products are then stabilized by SGS3, copied into dsRNAs by RDR6, sequentially processed by DCL4 and loaded into AGO1, resulting in target mRNA degradation.Citation9,Citation10
In recent years, most publications regarding ta-siRNAs biogenesis centered on discovery of ta-siRNA formation reasons and a plenty of mechanisms have been raised, such as “two-hit” models, “22-nt miRNAs as specific ta-siRNA initiators,” “asymmertrical bulged bases between miRNAs and miRNA duplex as sufficient structures for ta-siRNA transitivity,” however, SGS3-RDR6-DCL4, which was found nearly a decade years ago, are still considered as the sole pathway for ta-siRNA biogenesis.Citation11-Citation13 Moreover, although genome-wide DNA methylome sequencing studies detected cytosine methylation occurred in TAS gene loci, the reasons and mechanisms are largely unknown.Citation14
In recent work, though bisulfite sequencing analysis, we detected high cytosine DNA methylation confined at regions close to the miRNA cleavage site of TAS genes, suggesting that ta-siRNAs may have alternative actions that trigger DNA methylation for transcriptional regulation. As expected, SGS3 and RDR6, the key players for ta-siRNA formation, are found indispensable for TAS-derived siRNA directed DNA methylation. However, another important component for ta-siRNA generation, DCL4, is not required during this process.Citation15 This unexpected result provides an opportunity for us to test whether other DCL proteins involve in ta-siRNA biogenesis. Although it has been reported that DCL2, DCL3 and DCL4 can function redundantly in ta-siRNA production,Citation16 our further studies showed DNA methylation at TAS3 locus still persists in dcl2, dcl3, dcl4 double and triple mutants. Since DCL1 is also required for miRNA production, previous research cannot define the dicing role of DCL1 to TAS transcript is direct or indirect. Hence, we first determined the exact role of DCL1 in the process of ta-siRNA production: Besides miRNA formation, DCL1 can directly process TAS transcripts to produce TAS siRNAs for mediation of DNA methylation. These findings indicate both DCL1 and DCL4 can process TAS gene dsRNA transcripts.Citation15
Another important question is that ta-siRNAs interact with which AGO proteins to trigger DNA methylation. High-throughput sequencing results have revealed that most of ta-siRNAs from TAS loci were sorted into AGO1 and AGO2,Citation17 however, cytosine methylation triggered by ta-siRNAs are not compromised in ago1 and ago2 mutants.Citation15 Although only a small number of TAS siRNA are loaded into AGO4 and AGO6, our genetic analyses found that the interaction of AGO4 clade proteins and ta-siRNAs are sufficient to trigger epigenetic regulation. These results suggest besides of the 5′ terminal nucleotide and their biogenesis pathway, sRNAs loading into AGOs is also determined by their functions to some extent.Citation7,Citation15,Citation17 There may be a signal that can connect with sRNAs subsequent loading and ongoing functions.
In plants, silencing at endogenous transposons and repeats loci is usually guided by homologous 24-nt hc-siRNAs via RNA-directed DNA methylation (RdDM) pathway.Citation18 However, 21-nt ta-siRNA guided DNA methylation are not dependent on upstream RdDM pathway players (Pol IV, DCL3, RDR2), but strictly relied on downstream players (AGO4, Pol V, and so on), suggesting that each type of siRNA triggered DNA methylation utilize the same effector counterparts in RdDM pathway.Citation15
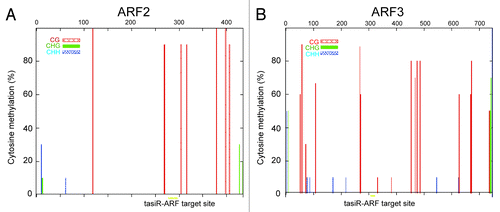
Although we unveiled the exact biogenesis and effector players for ta-siRNA mediated DNA methylation pathway, however, the precise biological role of this methylation remains obscure because no change of TAS gene expression took place when the methylation level was dramatically reduced.Citation15 Moreover, we detected the DNA methylation level of TAS3 ta-siRNA targets (ARF2 and ARF3) loci, but neither CHG nor CHH methylation were found around the target sites ( and ). Therefore, other investigations in the coming years are essential to explore precise modulation roles of ta-siRNA guided DNA methylation.
Figure 1.DNA methylation level at TAS3 ta-siRNA target gene loci. A, Analysis of DNA methylation at ARF2 gene locus by bisulfite sequencing in Col-0 plants .B, ARF3 locus DNA methylation. Sequencing data were analyzed by using Kismeth softwareCitation19 (http://katahdin.cshl.edu/homepage/kismeth/revpage.pl). The colored lines above the x axis show the percent methylation at individual cytosine sites. The positions corresponding to the tasiR-ARF sites are designated by grey bars (yellow bars in the online version).
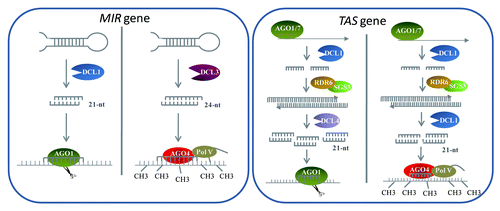
In conclusion, our here publication of DCL1-AGO4-DNA methylation for ta-siRNAs and previous work on DCL3-AGO4-DNA methylation for miRNAs indicate the existence of much complex non-canonical small RNA biogenesis and action pathways in plants ().
Figure 2. Schematic model for complex biogenesis and action pathway of miRNAs and ta-siRNAs. Two distinct pathways for miRNAs and ta-siRNAs that operate at mRNA and chromatin levels are shown. In the left box, 21-nt miRNAs are typically produced by DCL1 and loaded into AGO1 to direct target genes mRNA cleavage or protein translation inhibition. Alternatively, a class of 24-nt miRNAs are generated by DCL3 and recruited into AGO4 to guide DNA methylation transcriptional regulation. In the right box, TAS gene transcripts are initiated by miRNA-AGO1/7 interacting cleavage complexes and then stabilized and converted into dsRNA by SGS3 and RDR6. Most of dsRNAs are processed by DCL4 to generate 21-nt ta-siRNAs associating with AGO1 for targets posttranscriptional modulation. Meanwhile, some of dsRNAs are processed by DCL1 to produce siRNAs interacting with AGO4 clade proteins and Pol V-scaffold RNA complexes for trigging epigenetic regulation though DNA methylation.
Acknowledgments
This work was supported by the the National “863” program of Ministry of Science and Technology of China (#2012AA10A308), the National Transgenic Key Project (#2011ZX08009–001), and a grant from the Beijing Municipal Government.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat Rev Genet 2009; 10:94 - 108; http://dx.doi.org/10.1038/nrg2504; PMID: 19148191
- Voinnet O. Origin, biogenesis, and activity of plant microRNAs. Cell 2009; 136:669 - 87; http://dx.doi.org/10.1016/j.cell.2009.01.046; PMID: 19239888
- Golden TA, Schauer SE, Lang JD, Pien S, Mushegian AR, Grossniklaus U, et al. SHORT INTEGUMENTS1/SUSPENSOR1/CARPEL FACTORY, a Dicer homolog, is a maternal effect gene required for embryo development in Arabidopsis. Plant Physiol 2002; 130:808 - 22; http://dx.doi.org/10.1104/pp.003491; PMID: 12376646
- Xie Z, Kasschau KD, Carrington JC. Negative feedback regulation of Dicer-Like1 in Arabidopsis by microRNA-guided mRNA degradation. Curr Biol 2003; 13:784 - 9; http://dx.doi.org/10.1016/S0960-9822(03)00281-1; PMID: 12725739
- Brodersen P, Sakvarelidze-Achard L, Bruun-Rasmussen M, Dunoyer P, Yamamoto YY, Sieburth L, et al. Widespread translational inhibition by plant miRNAs and siRNAs. Science 2008; 320:1185 - 90; http://dx.doi.org/10.1126/science.1159151; PMID: 18483398
- Rajagopalan R, Vaucheret H, Trejo J, Bartel DP. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana.. Genes Dev 2006; 20:3407 - 25; http://dx.doi.org/10.1101/gad.1476406; PMID: 17182867
- Wu L, Zhou H, Zhang Q, Zhang J, Ni F, Liu C, et al. DNA methylation mediated by a microRNA pathway. Mol Cell 2010; 38:465 - 75; http://dx.doi.org/10.1016/j.molcel.2010.03.008; PMID: 20381393
- Allen E, Xie Z, Gustafson AM, Carrington JC. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 2005; 121:207 - 21; http://dx.doi.org/10.1016/j.cell.2005.04.004; PMID: 15851028
- Yoshikawa M, Peragine A, Park MY, Poethig RS. A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev 2005; 19:2164 - 75; http://dx.doi.org/10.1101/gad.1352605; PMID: 16131612
- Vazquez F, Vaucheret H, Rajagopalan R, Lepers C, Gasciolli V, Mallory AC, et al. Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol Cell 2004; 16:69 - 79; http://dx.doi.org/10.1016/j.molcel.2004.09.028; PMID: 15469823
- Axtell MJ, Jan C, Rajagopalan R, Bartel DP. A two-hit trigger for siRNA biogenesis in plants. Cell 2006; 127:565 - 77; http://dx.doi.org/10.1016/j.cell.2006.09.032; PMID: 17081978
- Chen HM, Chen LT, Patel K, Li YH, Baulcombe DC, Wu SH. 22-Nucleotide RNAs trigger secondary siRNA biogenesis in plants. Proc Natl Acad Sci USA 2010; 107:15269 - 74; http://dx.doi.org/10.1073/pnas.1001738107; PMID: 20643946
- Manavella PA, Koenig D, Weigel D. Plant secondary siRNA production determined by microRNA-duplex structure. Proc Natl Acad Sci USA 2012; 109:2461 - 6; http://dx.doi.org/10.1073/pnas.1200169109; PMID: 22308502
- Lister R, O’Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, et al. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 2008; 133:523 - 36; http://dx.doi.org/10.1016/j.cell.2008.03.029; PMID: 18423832
- Wu L, Mao L, Qi Y. Roles of DICER-LIKE and ARGONAUTE proteins in TAS-derived siRNAs triggered DNA methylation. Plant Physiol 2012; 160:990 - 9; http://dx.doi.org/10.1104/pp.112.200279; PMID: 22846193
- Henderson IR, Zhang X, Lu C, Johnson L, Meyers BC, Green PJ, et al. Dissecting Arabidopsis thaliana DICER function in small RNA processing, gene silencing and DNA methylation patterning. Nat Genet 2006; 38:721 - 5; http://dx.doi.org/10.1038/ng1804; PMID: 16699516
- Mi S, Cai T, Hu Y, Chen Y, Hodges E, Ni F, et al. Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell 2008; 133:116 - 27; http://dx.doi.org/10.1016/j.cell.2008.02.034; PMID: 18342361
- Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet 2010; 11:204 - 20; http://dx.doi.org/10.1038/nrg2719; PMID: 20142834
- Gruntman E, Qi Y, Slotkin RK, Roeder T, Martienssen RA, Sachidanandam R. Kismeth: analyzer of plant methylation states through bisulfite sequencing. BMC Bioinformatics 2008; 9:371; http://dx.doi.org/10.1186/1471-2105-9-371; PMID: 18786255