Abstract
The Arabidopsis trehalose-6-phosphate phosphatase (TPP) gene family arose mainly from whole genome duplication events and consists of 10 genes (TPPA-J). All the members encode active TPP enzymes, possibly regulating the levels of trehalose-6-phosphate, an established signaling metabolite in plants. GUS activity studies revealed tissue-, cell- and stage-specific expression patterns for the different members of the TPP gene family. Here we list additional examples of the remarkable features of the TPP gene family. TPPA-J expression levels seem, in most of the cases, differently regulated in response to light, darkness and externally supplied sucrose. Disruption of the TPPB gene leads to Arabidopsis plants with larger leaves, which is the result of an increased cell number in the leaves. Arabidopsis TPPA and TPPG are preferentially expressed in atrichoblast cells. TPPA and TPPG might fulfill redundant roles during the differentiation process of root epidermal cells, since the tppa tppg double mutant displays a hairy root phenotype, while the respective single knockouts have a distribution of trichoblast and atrichoblast cells similar to the wild type. These new data portray redundant and non-redundant functions of the TPP proteins in regulatory pathways of Arabidopsis.
Trehalose is a common disaccharide in bacteria, fungi and invertebrates, where it serves as a carbon source, structural component and stress protectant.Citation1 The most widespread trehalose biosynthesis pathway consists of two enzymatic reactions mediated by trehalose-6-phosphate synthase (TPS) and trehalose-6-phosphate phosphatase (TPP).Citation2 Although the amounts of trehalose and its intermediate trehalose-6-phosphate (T6P) are very scarce in higher vascular plants, plant genomes do contain multiple TPS and TPP genes. In Arabidopsis thaliana, trehalose biosynthesis genes are classified in three subfamilies based on their sequence homology to microbial TPS and TPP genes.Citation3-Citation6 Class I genes (AtTPS1–4) display the highest similarity to the yeast TPS, with AtTPS1 encoding the only active TPS enzyme.Citation7,Citation8 Class II genes (AtTPS5-AtTPS11) are more similar to the yeast TPP, but do not seem to encode any active TPS or TPP enzyme.Citation9-Citation11 The TPP family members (AtTPPA-J) have the conserved phosphatase boxes, typical for TPP enzymes.Citation12 The ancestor of the TPP family has probably been acquired by horizontal gene transfer from certain archaea or bacteriaCitation13 and recently, its members were shown to encode functional TPP enzymes in Arabidopsis.Citation14
Introducing heterologous trehalose biosynthesis genes in plants leads to increased stress tolerance and altered growth and morphology, while knocking out trehalose biosynthesis genes results in embryo-lethality and irregular branching of inflorescences.Citation3,Citation14-Citation18 Opposite phenotypes were obtained when either TPS or TPP enzymes were introduced in plants, pointing to an important role for T6P, the intermediate molecule in the biosynthesis pathway. It is now clear that T6P is an important signaling molecule, regulating the carbon status and starch biosynthesis in plants.Citation5,Citation19,Citation20
We showed in a recent study with the aid of a collinearity analysis that the Arabidopsis TPP gene family mainly originated from whole genome duplications. TPP activity was assayed for the 10 TPP proteins using a complementation assay in yeast. All the TPP members can be considered active TPP enzymes since they restore growth of the yeast tps2∆ strain at elevated temperature. Promoter-GUS studies revealed tissue-, cell- and stage-specific expression patterns for each of the TPP genes, indicating that TPP proteins may fulfill important regulatory functions by locally controlling T6P levels. Moreover, the functional diversity of the TPP family in Arabidopsis was demonstrated by the altered ABA-sensitivity of the tppg mutant.Citation14
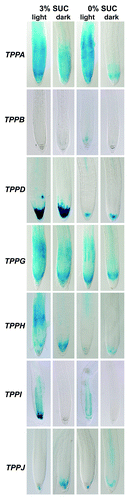
Plants use the metabolite T6P to signal their sugar status and to regulate their carbon use.Citation5,Citation20,Citation21 As such, T6P levels in the different plant organs, tissues and cell types, and upon environmental changes have to be tightly controlled. Here, we investigated whether a varying sugar supply and/or light program affect TPP gene expression in Arabidopsis seedlings. Therefore, promoter TPP::GUS-GFP lines were grown for 7 d on standard MS culture plates (described in ref. Citation14), supplemented with 0% and 3% sucrose and subsequently kept in the dark or continuous light for 3 d. GUS stained seedlings showed that starvation conditions (darkness and 0% sugar) led to a downregulation of TPPA, TPPD, TPPG, TPPH and TPPI expression in the root tip (). The absence of light seemed to have a bigger impact on TPPA and TPPI expression than a lack in sucrose (), while the opposite was noticed for TPPD expression (). Interestingly, the root expression pattern of TPPA in the absence/presence of light and sugars is highly similar to the one of TPPG, whereas the remaining, above-mentioned TPPs display distinct root expression profiles. TPPC, TPPE and TPPF genes are not expressed in expressed in roots.Citation14 The variable GUS staining patterns observed in the root tips of promoter TPP::GUS-GFP lines suggest a subtle, mostly unique regulation of TPP gene expression in response to altered sugar availability and light conditions. These findings are indications that the 10 TPP enzymes in Arabidopsis could function in local networks, integrating environmental signals with metabolic processes, through the breakdown of T6P.
Figure 1. Histochemical localization of GUS activity in root tips of promoter TPP::GUS-GFP lines under different sugar and light conditions. 7-d-old seedlings were grown on MS media supplemented with 0% and 3% sucrose (SUC) and kept for three additional days in continuous light or dark before sampling.
TPPs are known to influence plant growth and development. We found that altering the gene expression of one of the plant endogenous TPPs, TPPB, affects the shoot size of Arabidopsis plants. Disrupting the TPPB gene leads to a significant increase in leaf area, as seen in the tppb-1 knockout (SALK_037324,Citation14,Citation22) and the tppb-2 knockdown (Sail_191F08,Citation14,Citation23) after a growth period of 21 d on MS culture plates (). TPPB-1 and TPPB-2 overexpressing Arabidopsis plantsCitation14 showed the opposite phenotype in vitro (). In addition, tppb mutants display on average 2 more leaves than Wt plants, whereas TPPB overexpressors seem delayed in growth (). TPPB is expressed in the shoot apical meristem and at the basal end of young, expanding leaves.Citation14 This expression profile resembles to the one in developing leaves of pCYCB1;1-D-box:GUS lines used for tracking the cell proliferation zone, which gradually disappears from the leaf tip on.Citation24 These expression profiles suggest that TPPB rather interferes with leaf growth during early leaf development than during the maturation stages. Since the initial growth phase in leaves is mostly driven by cell divisions,Citation25 we investigated whether the tppb leaf phenotype was due to an increased cell number or due to a larger epidermal cell area. The first pair of true leaves from 21-d-old mutants, grown on MS culture plates, were prepared for microscopic analysis as described in De Veylder et al.Citation26 Images were taken from basal and apical sections of the abaxial epidermis and analyzed by imageJ. Leaves of the tppb-1 and tppb-2 mutants were 17 to 30% larger than the ones from the Wt (). The estimated cell number per leaf was significantly higher in the tppb mutants, displaying 13% to 37% more cells than the Wt, while the epidermal cell size seemed unaffected (). Since the majority of pavement cells originate from asymmetric divisions of the stomatal lineage,Citation27 stomatal indexes were determined as well, but no significant differences were observed (). In contrast with the tppb knockouts, the leaf area of TPPB-1 plants was reduced by 22% compared with the Wt, while the leaf area of the TPPB-2 line seemed indistinguishable from Wt Col-0 in this assay (). The reduced leaf size of the TPPB-1 overexpressor seemed attributed to a decrease of 13% in the estimated cell number (). These data indicate that the increased shoot size of the tppb mutants could be the result of higher cell division rates or due to a delay in the initiation of cell differentiation.Citation28 In opposite, a lower cell division frequency or an earlier onset of cell expansion might have caused the presence of smaller leaves in the TPPB-1 overexpressing line. A possible role for the trehalose metabolism in the cell cycle regulation of Arabidopsis plants has been suggested earlier. TPS1, the only enzyme that catalyzes the synthesis of T6P, is known to interact with the cell cycle-dependent kinase CKA;1, the kinesin KCA;1 and β-tubulin.Citation29 In silico predictions (http://suba.plantenergy.uwa.edu.au/; refs. 30 and 31), and microarray data sets seem to further support a link between trehalose metabolism and cell cycle regulation as there is co-expression between the TPPB gene, the kinesin-encoding ATK5 gene and AUR1, encoding a kinase which associates with the spindle and cytoskeletal structures necessary for cytokinesis.Citation32 Moreover, embryos of the tps1 mutant show a decreased cell division rate, which could be the result of impaired trehalose signaling.Citation33 Alternatively, the trehalose metabolism might, as an integrator of the nutritional status and growth, influence the regulation of cell division.Citation34 The exact role(s) of TPPB and the trehalose metabolism in the cell cycle regulation still remains elusive.
Figure 2. Shoot phenotype of Wt Col-0 and TPPB mutant plants grown for 21 d on MS culture plates. (A) Rosettes of tppb-1 and tppb-2 mutants are significantly bigger than Wt Col-0 rosettes, at p < 0.001 (Student’s t-test, n = 6–10). In opposite, TPPB-1 and TPPB-2 overexpressors display significantly smaller rosettes compared with Wt Col-0, at p < 0.05 (Student’s t-test, n = 6–10). Numbers indicate the total leaf size ± SD (B) Area of the individual rosette leaves shown in (A): Wt Col-0 (green bars), tppb-1 (blue line), tppb-2 (red line), TPPB-1 (black line) and TPPB-2 (orange line). Error bars represent averages ± SD (n = 6–10).
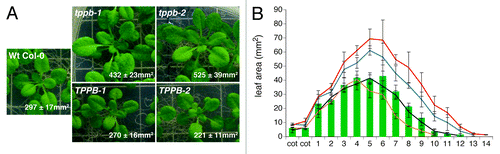
Table 1. Abaxial epidermal analysis of the first pair of true leaves in 21-d-old TPPB mutants
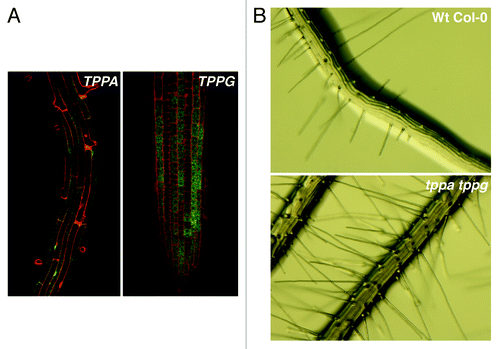
Another example that demonstrates the importance of TPP proteins during plant development, is the hairy root phenotype of the tppa tppg double mutant. The root epidermis of Arabidopsis Wt plants contains files with trichoblasts (root hair-producing cells) and files with atrichoblasts (hairless cells). These two types of files are located at distinct positions within the root, which implies that the fate of root epidermal cells depends on the location rather than the lineage.Citation35 When analyzing roots of the promoter TPP::GUS-GFP lines (described in ref. Citation14), TPPA and TPPG expression was seen in the atrichoblast cells of the root elongation and meristematic zones, respectively (). These expression patterns suggest that local TPP transcription might be important for proper development of atrichoblast cells. When looking at the single tppa (GABI_016E11,Citation14,Citation36) and tppg (Salk_078443,Citation14,Citation22) knockout mutants, the root epidermal cells were distributed similarly to the Wt with alternate trichoblast and atrichoblast files (data not shown). However, in the tppa tppg double mutant (), the root epidermis consists of trichoblast files at some atrichoblast positions. This finding suggests that the presence of TPPA and TPPG promotes atrichoblast fate over trichoblast fate in a redundant way. A possible redundancy in function of these TPPs is supported by the fact that TPPA and TPPG are clustered together in the phylogenetic tree of plant TPPs.Citation14 Furthermore, an Arabidopsis microarray study showed that TPPA and TPPG are co-expressed with a variety of genes involved in processes that trigger tip growth, such as vesicle-mediated transport, cellular membrane fusion, cell wall biogenesis, rearrangement of the cytoskeleton and calcium-dependent signaling.Citation32,Citation37 It seems thus likely that the TPPA and TPPG enzymes are possible inhibitors of the cellular processes associated with root hair tip growth. This assumption must however be taken with care since the differentiation program of root epidermal cells is very complex and starts early on, prior to the outgrowth of the root hair itself.Citation37
Figure 3. The role of TPPA and TPPG during the development of root epidermal cells in Arabidopsis seedlings. (A) Promoter TPP::GUS-GFP lines show TPPA and TPPG expression in atrichoblasts of root elongation and root meristematic zones, respectively. (B) The root of Wt Col-0 seedlings consists of files with trichoblasts and atrichoblasts, while the root epidermis of the tppa tppg double mutant is restricted to files with trichoblasts.
We can conclude that the 10 members of the TPP gene family are not only specifically expressed across the plant and during the different stages of development, but most of their expression levels seem also to differ in response to a changing environment. Disruption of the TPPB gene leads to plants with larger leaves, which seems the result of an increased amount of epidermal cells. TPPA and TPPG proteins on the other hand, likely fulfill a redundant role during the development of atrichoblasts in the root epidermis. Altogether, these observations illustrate the non-redundancy and redundancy in function of the large TPP multigene family in Arabidopsis thaliana.
Acknowledgments
We thank Nico Van Goethem for the assistance with preparation of the figures and Dr. Frantisek Baluska for the invitation to write this short communication. This research was supported by grants from the Fund for Scientific Research Flanders (FWO G.0859.10), the Industrial Research Fund of the KU Leuven (IOF/KP/08/001) and the VIB International PhD Program 2006.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Elbein AD, Pan YT, Pastuszak I, Carroll D. New insights on trehalose: a multifunctional molecule. Glycobiology 2003; 13:17R - 27R; http://dx.doi.org/10.1093/glycob/cwg047; PMID: 12626396
- Cabib E, Leloir LF. The biosynthesis of trehalose phosphate. J Biol Chem 1958; 231:259 - 75; PMID: 13538966
- Goddijn OJM, Verwoerd TC, Voogd E, Krutwagen RWHH, de Graaf PTHM, van Dun K, et al. Inhibition of trehalase activity enhances trehalose accumulation in transgenic plants. Plant Physiol 1997; 113:181 - 90; http://dx.doi.org/10.1104/pp.113.1.181; PMID: 9008394
- Leyman B, Van Dijck P, Thevelein JM. An unexpected plethora of trehalose biosynthesis genes in Arabidopsis thaliana.. Trends Plant Sci 2001; 6:510 - 3; http://dx.doi.org/10.1016/S1360-1385(01)02125-2; PMID: 11701378
- Lunn JE, Feil R, Hendriks JHM, Gibon Y, Morcuende R, Osuna D, et al. Sugar-induced increases in trehalose 6-phosphate are correlated with redox activation of ADPglucose pyrophosphorylase and higher rates of starch synthesis in Arabidopsis thaliana.. Biochem J 2006; 397:139 - 48; http://dx.doi.org/10.1042/BJ20060083; PMID: 16551270
- Lunn JE. Gene families and evolution of trehalose metabolism in plants. Funct Plant Biol 2007; 34:550 - 63; http://dx.doi.org/10.1071/FP06315
- Blázquez MA, Santos E, Flores CL, Martínez-Zapater JM, Salinas J, Gancedo C. Isolation and molecular characterization of the Arabidopsis TPS1 gene, encoding trehalose-6-phosphate synthase. Plant J 1998; 13:685 - 9; http://dx.doi.org/10.1046/j.1365-313X.1998.00063.x; PMID: 9681010
- Vandesteene L, Ramon M, Le Roy K, Van Dijck P, Rolland F. A single active trehalose-6-P synthase (TPS) and a family of putative regulatory TPS-like proteins in Arabidopsis. Mol Plant 2010; 3:406 - 19; http://dx.doi.org/10.1093/mp/ssp114; PMID: 20100798
- Vogel G, Fiehn O, Jean-Richard-dit-Bressel L, Boller T, Wiemken A, Aeschbacher RA, et al. Trehalose metabolism in Arabidopsis: occurrence of trehalose and molecular cloning and characterization of trehalose-6-phosphate synthase homologues. J Exp Bot 2001; 52:1817 - 26; http://dx.doi.org/10.1093/jexbot/52.362.1817; PMID: 11520870
- Harthill JE, Meek SE, Morrice N, Peggie MW, Borch J, Wong BH, et al. Phosphorylation and 14-3-3 binding of Arabidopsis trehalose-phosphate synthase 5 in response to 2-deoxyglucose. Plant J 2006; 47:211 - 23; http://dx.doi.org/10.1111/j.1365-313X.2006.02780.x; PMID: 16771775
- Ramon M, De Smet I, Vandesteene L, Naudts M, Leyman B, Van Dijck P, et al. Extensive expression regulation and lack of heterologous enzymatic activity of the Class II trehalose metabolism proteins from Arabidopsis thaliana.. Plant Cell Environ 2009; 32:1015 - 32; http://dx.doi.org/10.1111/j.1365-3040.2009.01985.x; PMID: 19344332
- Thaller MC, Schippa S, Rossolini GM. Conserved sequence motifs among bacterial, eukaryotic, and archaeal phosphatases that define a new phosphohydrolase superfamily. Protein Sci 1998; 7:1647 - 52; http://dx.doi.org/10.1002/pro.5560070722; PMID: 9684901
- Avonce N, Wuyts J, Verschooten K, Vandesteene L, Van Dijck P. The Cytophaga hutchinsonii ChTPSP: First characterized bifunctional TPS-TPP protein as putative ancestor of all eukaryotic trehalose biosynthesis proteins. Mol Biol Evol 2010; 27:359 - 69; http://dx.doi.org/10.1093/molbev/msp241; PMID: 19812028
- Vandesteene L, López-Galvis L, Vanneste K, Feil R, Maere S, Lammens W, et al. Expansive evolution of the TREHALOSE-6-PHOSPHATE PHOSPHATASE gene family in Arabidopsis thaliana.. [epub ahead of print] Plant Physiol 2012; 160:884 - 96; http://dx.doi.org/10.1104/pp.112.201400; PMID: 22855938
- Holmström KO, Mäntylä E, Welin B, Mandal A, Palva ET, Tunnela OE, et al. Drought tolerance in tobacco. Nature 1996; 379:683 - 4; http://dx.doi.org/10.1038/379683a0
- Eastmond PJ, van Dijken AJH, Spielman M, Kerr A, Tissier AF, Dickinson HG, et al. Trehalose-6-phosphate synthase 1, which catalyses the first step in trehalose synthesis, is essential for Arabidopsis embryo maturation. Plant J 2002; 29:225 - 35; http://dx.doi.org/10.1046/j.1365-313x.2002.01220.x; PMID: 11851922
- Avonce N, Leyman B, Mascorro-Gallardo JO, Van Dijck P, Thevelein JM, Iturriaga G. The Arabidopsis trehalose-6-P synthase AtTPS1 gene is a regulator of glucose, abscisic acid, and stress signaling. Plant Physiol 2004; 136:3649 - 59; http://dx.doi.org/10.1104/pp.104.052084; PMID: 15516499
- Satoh-Nagasawa N, Nagasawa N, Malcomber S, Sakai H, Jackson D. A trehalose metabolic enzyme controls inflorescence architecture in maize. Nature 2006; 441:227 - 30; http://dx.doi.org/10.1038/nature04725; PMID: 16688177
- Paul MJ. Trehalose 6-phosphate: a signal of sucrose status. Biochem J 2008; 412:e1 - 2; http://dx.doi.org/10.1042/BJ20080598; PMID: 18426388
- Schluepmann H, Berke L, Sanchez-Perez GF. Metabolism control over growth: a case for trehalose-6-phosphate in plants. J Exp Bot 2012; 63:3379 - 90; http://dx.doi.org/10.1093/jxb/err311; PMID: 22058405
- Schluepmann H, van Dijken A, Aghdasi M, Wobbes B, Paul M, Smeekens S. Trehalose mediated growth inhibition of Arabidopsis seedlings is due to trehalose-6-phosphate accumulation. Plant Physiol 2004; 135:879 - 90; http://dx.doi.org/10.1104/pp.104.039503; PMID: 15181209
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana.. Science 2003; 301:653 - 7; http://dx.doi.org/10.1126/science.1086391; PMID: 12893945
- Sessions A, Burke E, Presting G, Aux G, McElver J, Patton D, et al. A high-throughput Arabidopsis reverse genetics system. Plant Cell 2002; 14:2985 - 94; http://dx.doi.org/10.1105/tpc.004630; PMID: 12468722
- Eloy NB, de Freitas Lima M, Van Damme D, Vanhaeren H, Gonzalez N, De Milde L, et al. The APC/C subunit 10 plays an essential role in cell proliferation during leaf development. Plant J 2011; 68:351 - 63; http://dx.doi.org/10.1111/j.1365-313X.2011.04691.x; PMID: 21711400
- Donnelly PM, Bonetta D, Tsukaya H, Dengler RE, Dengler NG. Cell cycling and cell enlargement in developing leaves of Arabidopsis. Dev Biol 1999; 215:407 - 19; http://dx.doi.org/10.1006/dbio.1999.9443; PMID: 10545247
- De Veylder L, Beeckman T, Beemster GT, de Almeida Engler J, Ormenese S, Maes S, et al. Control of proliferation, endoreduplication and differentiation by the Arabidopsis E2Fa-DPa transcription factor. EMBO J 2002; 21:1360 - 8; http://dx.doi.org/10.1093/emboj/21.6.1360; PMID: 11889041
- Bergmann DC, Sack FD. Stomatal development. Annu Rev Plant Biol 2007; 58:163 - 81; http://dx.doi.org/10.1146/annurev.arplant.58.032806.104023; PMID: 17201685
- Beemster GT, De Veylder L, Vercruysse S, West G, Rombaut D, Van Hummelen P, et al. Genome-wide analysis of gene expression profiles associated with cell cycle transitions in growing organs of Arabidopsis. Plant Physiol 2005; 138:734 - 43; http://dx.doi.org/10.1104/pp.104.053884; PMID: 15863702
- Geelen D, Royackers K, Vanstraelen M, Inze D, Van Dijck P, Thevelein JM, et al. Trehalose-6-P synthase: AtTPS1 high molecular weight complexes in yeast and Arabidopsis. Plant Sci 2007; 173:426 - 37; http://dx.doi.org/10.1016/j.plantsci.2007.07.002
- Heazlewood JL, Tonti-Filippini J, Verboom RE, Millar AH. Combining experimental and predicted datasets for determination of the subcellular location of proteins in Arabidopsis.. Plant Physiol 2005; 139:598 - 609; http://dx.doi.org/10.1104/pp.105.065532; PMID: 16219920
- Heazlewood JL, Verboom RE, Tonti-Filippini J, Small I, Millar AH. SUBA: the Arabidopsis subcellular database. Nucleic Acids Res 2007; 35:Database issue D213 - 8; http://dx.doi.org/10.1093/nar/gkl863; PMID: 17071959
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol 2004; 136:2621 - 32; http://dx.doi.org/10.1104/pp.104.046367; PMID: 15375207
- Gómez LD, Baud S, Gilday A, Li Y, Graham IA. Delayed embryo development in the ARABIDOPSIS TREHALOSE-6-PHOSPHATE SYNTHASE 1 mutant is associated with altered cell wall structure, decreased cell division and starch accumulation. Plant J 2006; 46:69 - 84; http://dx.doi.org/10.1111/j.1365-313X.2006.02662.x; PMID: 16553896
- Riou-Khamlichi C, Menges M, Healy JM, Murray JA. Sugar control of the plant cell cycle: differential regulation of Arabidopsis D-type cyclin gene expression. Mol Cell Biol 2000; 20:4513 - 21; http://dx.doi.org/10.1128/MCB.20.13.4513-4521.2000; PMID: 10848578
- Masucci JD, Rerie WG, Foreman DR, Zhang M, Galway ME, Marks MD, et al. The homeobox gene GLABRA2 is required for position-dependent cell differentiation in the root epidermis of Arabidopsis thaliana.. Development 1996; 122:1253 - 60; PMID: 8620852
- Rosso MG, Li Y, Strizhov N, Reiss B, Dekker K, Weisshaar B. An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol Biol 2003; 53:247 - 59; http://dx.doi.org/10.1023/B:PLAN.0000009297.37235.4a; PMID: 14756321
- Ryan E, Steer M, Dolan L. Cell biology and genetics of root hair formation in Arabidopsis thaliana.. Protoplasma 2001; 215:140 - 9; http://dx.doi.org/10.1007/BF01280310; PMID: 11732053