 ?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
Following the discovery that in Arabidopsis, a third isoenzyme of NADH-dependent glutamate dehydrogenase (GDH) is expressed in the mitochondria of the root companion cells, we have re-examined the GDH isoenzyme composition. By analyzing the NADH-GDH isoenzyme composition of single, double and triple mutants deficient in the expression of the three genes encoding the enzyme, we have found that the α, β and γ polypeptides that comprise the enzyme can be assembled into a complex combination of heterohexamers in roots. Moreover, we observed that when one or two of the three root isoenzymes were missing from the mutants, the remaining isoenzymes compensated for this deficiency. The significance of such complexity is discussed in relation to the metabolic and signaling function of the NADH-GDH enzyme. Although it has been shown that a fourth gene encoding a NADPH-dependent enzyme is present in Arabidopsis, we were not able to detect corresponding enzyme activity, even in the triple mutant totally lacking NADH-GDH activity.
In the last 25 years, good progress has been made in obtaining a better understanding of the role of alternative metabolic pathways in plants, which are potentially able to incorporate ammonium into organic molecules.Citation1 The reaction catalyzed by the mitochondrial enzyme NADH-dependent glutamate dehydrogenase (NADH-GDH, EC 1.4.1.2), has often been considered as being one of such alternative pathways, as the enzyme is able to incorporate ammonium into 2-oxoglutarate to form glutamate in vitro, or to function in the opposite direction to deaminate glutamate. Although it has been clearly demonstrated by the means of 15N- and 13C-labeling experiments that the latter deamination reaction occurs in plant cells, it has been regularly argued that under certain physiological conditions, when the ammonium concentration reaches a particular threshold, the enzyme is able to function in the aminating direction.Citation2-Citation5 Consequently, the exact physiological function of the enzyme NADH-GDH was still matter of debate until Fontaine et al.,Citation6 demonstrated that in Arabidopsis thaliana three distinct nuclear genes each encode an active GDH protein. One named GDH3 is expressed only in the root companion cells, whereas the two others GDH1 and GDH2 are expressed in the same companion cells in both roots and shoots. The finding that a third NADH-GDH isoenzyme is active in Arabidopsis has demonstrated through the use of mutants deficient in the expression of the three NADH-GDH genes, that the main function of the enzyme is to provide 2-oxoglutatarate for the tricarboxylic acid cycle, primarily in the roots.Citation6 Thus, such a demonstration reinforced the current consensus that NADH-GDH plays virtually no role in the assimilation of inorganic nitrogen and that, whatever its metabolic origin, ammonia is mostly if not exclusively incorporated into organic molecules through the glutamine synthetase-glutamate synthase (GS-GOGAT) assimilatory pathway.Citation7,Citation8
Prior to the discovery of a third active GDH isoenzyme in roots, it was considered that in most plant species, GDH is encoded by two distinct nuclear genes.Citation4,Citation9,Citation10 GDH1 gene encodes a polypeptide subunit termed β and GDH2 gene encodes α and the polypeptides can be assembled as homo- or heterohexamers composed of different ratios of α and β, leading to the formation of seven active isoenzymes. These seven isoenzymes can be separated by native PAGE (PAGE), followed by in-gel staining for NAD-dependent GDH activityCitation11,Citation12 or NADH-dependent GDH activity,Citation13 thus indicating that GDH exhibits a similar anabolic and catabolic activity in vitro. In addition, variations in the GDH isoenzyme pattern were observed according to the organ examined, or the N source supplied to the plant.Citation14,Citation15 However, the physiological functions of the seven GDH isoenzymes and the significance of the variations in their relative proportions in roots and shoots, remained unknown.
The Presence of a Third Active NADH-GDH Polypeptide in Roots Leads to the Formation of a Complex Pattern of Isoenzymes
The finding of a third GDH gene encoding an isoenzyme termed γ that is only expressed in roots, prompted us to examine if the corresponding polypeptide could be assembled into heterohexamers with the previously indentified α- and β-subunits, which are expressed in both the roots and leaves. We have therefore re-examined the GDH isoenzyme content of the roots and leaves of the WT, the gdh1, gdh2 and gdh3 single mutants, the gdh1-2 double mutant and the gdh1–2-3 triple mutant, by staining for NAD-dependent GDH activity following separation by non-denaturing PAGE. For experimental details describing the production of the mutants and the in-gel staining for GDH activity (see refs. Citation6, Citation15 and Citation16). Following PAGE of extracts of roots of the wild type (WT), a faint eighth band of GDH enzyme activity was visible below the fastest moving, most anodal form of the standard seven GDH isoenzyme band pattern (). This additional band of NAD-GDH activity was designated isoenzyme γ.Citation6 For the gdh1 mutant, in which only the isoenzyme composed of α homohexamers remained, isoenzyme γ was just visible following PAGE, but at the limit of detection using in-gel activity staining ( and ). Both in the gdh2 and in the gdh1-2 mutant, the activity of isoenzyme γ was much higher compared with the WT ( and ). Following PAGE of extracts of roots of the gdh3 mutant, only the seven isoenzymes composed of homo- and heterohexamers of α and β were detected after activity staining,Citation6,Citation16 (see also ). As expected, in the gdh1-2-3 triple mutant, the eight bands of enzyme activity corresponding to the two α and β homohexamers, the five heterohexamers composed of different ratios of α and β polypeptides and the isoenzyme γ were not detected. Interestingly for the gdh2 mutant, seven isoenzymes were detected following PAGE of root extracts, thus suggesting that the β and γ polypeptides could be assembled into five heterohexamers. It is also likely that there were heterohexamers composed of different ratios of α and γ polypeptides both in the WT and the gdh1 mutant. However, the presence of α and γ heterohexamers could not be clearly distinguished using in-gel GDH activity staining, because the active isoenzymes containing the α and γ polypeptides ran very close to each other following PAGE. Nevertheless a smear of GDH activity, which could indicate the presence of isoenzyme bands, was visible in the most anodal section of the gel following PAGE and activity staining of extracts of the gdh1 mutant. This faint staining of anodal bands was also detected following PAGE of WT extracts and taken together suggests that the formation of heterohexamers between the α and γ polypeptides may have occurred (). Lastly, we observed a shift between the position of the seven isoenzyme bands visible in the WT or the gdh3 mutant and those present in the gdh2 mutant (). This shift can be explained by the formation of heterohexamers between the polypetides α, β and γ in the WT (listed on the right side of and indicated by successive arrows). The 28 possible isoenzyme combinations shown in agree with the formula provided by Shaw,Citation17 which assumes that there is random association of the subunits:
Figure 1. NAD-GDH isoenzyme patterns of roots of Arabidopsis wild type and mutants deficient in the three genes encoding the enzyme. (A) Protein extracts of the roots of the wild type (WT), gdh1, gdh2, gdh3, gdh1-2 and gdh1-2-3 mutants were subjected to native PAGE followed by NAD-GDH in-gel activity staining. The different subunit combinations of the eight isoenzymes detected in the WT are indicated on the left side of the panel. Double arrows indicate the shift observed between the gdh2 and the gdh3 mutants for the positions of the different isoenzymes. (B) Protein extracts of the roots of the WT, gdh1 and gdh2 mutants were subjected to native PAGE followed by NAD-GDH in-gel activity staining. The possible combinations of the three GDH subunits α, β and γ in the different isoenzymes, when they are all present in the WT are shown in the table at the right side of the panel. The subunit composition that predominates in the WT is indicated in red (black closed arrows). The red open arrows in the WT indicate the position of the other subunit composition also listed in the table on the right. The possible different combinations of the subunits in the GDH isoenzymes, when the subunit α or the subunit β is lacking in the gdh2 and gdh1 mutants respectively are also shown in blue (black closed arrows) and in the table on the right. The red open arrows in the gdh1 mutant indicate the position of the five heterohexamer isoenzymes and their composition is presented in the table on the right.
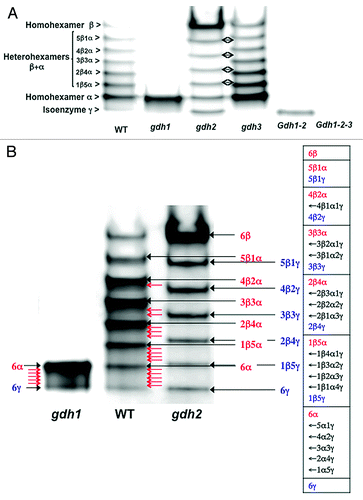
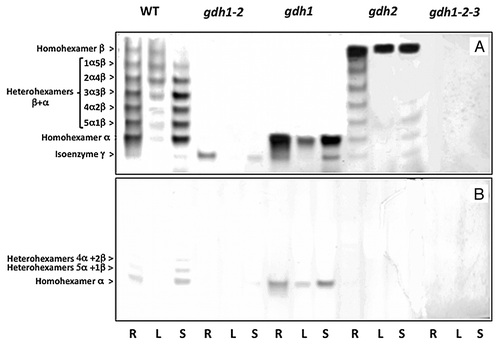
Figure 2. NADH- and NADPH-dependent GDH isoenzyme patterns of roots, leaves and floral stems of Arabidopsis WT and mutants deficient in the three genes encoding the enzyme. Protein extracts of the roots (R), leaves (L) and floral stems (S) of the wild type (WT), gdh1, gdh2, gdh1–2 and gdh1–2-3 mutants were subjected to native PAGE followed by NAD-GDH (A) and NADP-GDH (B) in-gel activity staining. The composition of the subunits of the eight isoenzymes in the WT are indicated on the left side of the panel.
Where i is the number of possible isoenzymes, P is the number of subunits (6) and S is the number of different types of subunits (3).
In the gdh2 mutant in which the isoenzyme α is not present, the possible formation of heterohexamers composed of different ratios of polypetides β and γ is also visible in as well as in .
It is also worth stressing that compensatory mechanisms for the isoenzyme composition of the roots appear to be occurring when one of the three isoenzymes is lacking. The in-gel staining for NAD-GDH activity presented in shows that compared with the WT, the activity of isoenzyme α was increased in the gdh1 mutant, whereas in the gdh2 mutant isoenzyme β was more active. An increase in the apparent activity of isoenzyme γ was also observed in the roots of the gdh2 and gdh1–2 mutant. Thus, it can be concluded that the compensatory mechanism, previously observed in the leaves of gdh1 and gdh2 mutants in which the activity of subunit α and β was increased respectively,Citation16 also occurs in roots. This compensation appears to occur when there is a deficiency in one of the subunits α, β or γ, that have the ability to be assembled into heterohexamers. The physiological significance of such a compensatory mechanism still remains to be elucidated. It may only be due to the fact that an optimal metabolic activity of the enzyme is required, or it may be a consequence of a more complex inter-organ signaling regulatory mechanism circulating via the phloem. Further studies are currently being performed to examine the impact of different combinations of single and double mutations on the metabolic phenotype of roots and shoots and phloem composition.
On the Occurrence of NADPH-Dependent GDH Activity
It is well established that in higher plants, the mitochondrial NAD(H)-dependent enzyme is by far the most active and is of universal occurrence. However in early work, NADP(H)-dependent GDH activity was found to be associated with plastids, although in some cases it was detected in the mitochondria.Citation18,Citation19 Later on, in a range of higher plants,Citation12,Citation20,Citation21 including Arabidopsis,Citation22,Citation23 the presence of low NADP-dependent GDH activity has regularly been reported. Since a fourth gene encoding a putative NADPH-GDH, has been identified in ArabidopsisCitation24,Citation25 and rice,Citation26 it was interesting to determine if NADP(H)-dependent activity could be detected in the different gdh mutants.
When the GDH isoenzyme content of extracts of different organs was examined in the WT following PAGE and staining for NAD-dependent GDH activity, eight bands were visible both in the roots and floral stems. However, following PAGE of leaf extracts, the standard seven bands isoenzyme pattern was observed. Although at the limit of detection following in-gel enzyme activity staining, we found that isoenzyme γ was also active in the floral stems. As previously observed in Arabidopsis,Citation15,Citation16 the most cathodal isoenzymes composed of homohexamers of β were more active in leaves, whereas the most anodal isoenzymes composed of homohexamer α were more active in both roots and floral stems (Fig.. 2). In leaves of the gdh1 mutant only the homohexamer composed of six α-subunits was detected. Compared with the WT, isoenzyme γ was more active in both the roots and stems of the gdh-1 mutant, following PAGE and in-gel activity staining. Only the homohexamer composed of six β-subunits was detected in the leaves of the gdh2-deficient mutant, whereas in both roots and stems, heterohexamers between the β and γ subunits were still present (see above). Following PAGE of an extract of the gdh1–2-3 mutant, no band of NAD-GDH enzyme activity was visible, whichever organ was examined ().
Interestingly, there would appear to be a compensatory mechanism for the activity of isoenzyme γ both in the gdh1,and in the gdh1–2 and to a lower extent in the gdh2 mutant mutants in comparison to the WT. As preciously observed,Citation6,Citation16 similar compensation mechanisms can be seen for homohexamer α and homohexamer β in gdh1 and gdh2 mutants respectively. Although the physiological significance of such mechanisms remains unresolved, one can hypothesize that the plant compensates for the lack of one of the three isoenzymes by overexpresing the remaining ones. This also suggests that there are metabolic signals controlling such regulatory mechanisms, thus highlighting the importance of the enzyme at the interface between carbon and nitrogen metabolism in both roots and shoots.
When NAD was replaced by NADP following PAGE, in-gel GDH activity staining revealed that in the WT only the bands composed of the α homohexamer and 5α-1β and 4α-2β heterohexamers exhibited very faint enzyme activity. Similarly in the gdh1 mutant, the remaining α homohexamer also exhibited NADP-dependent enzyme activity. Therefore, it can be concluded that only the α isoenzyme is able to use NADP as a co-factor. No additional band of NADP-dependent GDH activity could be detected following PAGE and in-gel staining of extracts of roots, leaves or stems. In particular, it can be seen that even in the GDH triple mutant gdh1–2-3, that is totally deficient of NAD-GDH activity, there is no evidence of any NADP dependent activity. Therefore, it would appear that the gene encoding a putative NADP(H)-GDH is not expressed as an active protein in Arabidopsis and that the NADP(H)-dependent enzyme activity detected by some authors may be due to the action of the α subunit of the NAD(H)-dependent GDH2 isoenzyme.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Joy KW. Ammonia, glutamine, and asparagine: a carbon-nitrogen interface. Can J Bot 1988; 66:2103 - 9
- Yamaya T, Oaks A. Synthesis of glutamate by mitochondria - an anaplerotic function for glutamate dehydrogenase. Physiol Plant 1987; 70:749 - 56; http://dx.doi.org/10.1111/j.1399-3054.1987.tb04334.x
- Oaks A. Evidence for deamination by glutamate dehydrogenase in higher plants. [reply] Can J Bot 1995; 73:1116 - 7; http://dx.doi.org/10.1139/b95-121
- Melo-Oliveria R, Cinha-Oliveria I, Coruzzi GM. Arabidopsis mutant analysis and gene regulation define a non-redundant role for glutamate dehydrogenase in nitrogen assimilation. Proc Natl Acad Sci USA 1996; 96:4718 - 23; http://dx.doi.org/10.1073/pnas.93.10.4718
- Skopelitis DS, Paranychianakis NV, Paschalidis KA, Pliakonis ED, Delis ID, Yakoumakis DI, et al. Abiotic stress generates ROS that signal expression of anionic glutamate dehydrogenases to form glutamate for proline synthesis in tobacco and grapevine. Plant Cell 2006; 18:2767 - 81; http://dx.doi.org/10.1105/tpc.105.038323; PMID: 17041150
- Fontaine JX, Tercé-Laforgue T, Armengaud P, Clément G, Renou JP, Pelletier S, et al. Characterization of a NADH-dependent glutamate dehydrogenase mutant of Arabidopsis demonstrates the key role of this enzyme in root carbon and nitrogen metabolism. Plant Cell 2012; 24:4044 - 65; http://dx.doi.org/10.1105/tpc.112.103689; PMID: 23054470
- Lea PJ, Miflin BJ. Nitrogen assimilation and its relevance to crop improvement. In: Foyer CH, Zhang H, eds, Nitrogen metabolism in plants in the post-genomic era. Chichester, UK: Wiley-Blackwell, 2011:1-40.
- Hirel B, Tétu T, Lea PJ, Dubois F. Improving nitrogen use efficiency in crops for sustainable agriculture. Sustainability 2011; 3:1452 - 85; http://dx.doi.org/10.3390/su3091452
- Restivo FM. Molecular cloning of glutamate dehydrogenase genes of Nicotiana plumbaginifolia: structure and regulation of their expression by physiological and stress conditions. Plant Sci 2004; 166:971 - 82; http://dx.doi.org/10.1016/j.plantsci.2003.12.011
- Purnell MP, Skopelitis DS, Roubelakis-Angelakis KA, Botella JR. Modulation of higher-plant NAD(H)-dependent glutamate dehydrogenase activity in transgenic tobacco via alteration of beta subunit levels. Planta 2005; 222:167 - 80; http://dx.doi.org/10.1007/s00425-005-1510-z; PMID: 15803323
- Thurman DA, Palin C, Laycock MV. Isoenzymatic nature of L-glutamic dehydrogenase of higher plants. Nature 1965; 207:193 - 4; http://dx.doi.org/10.1038/207193a0; PMID: 5886113
- Turano FJ, Dashner R, Upadhyaya A, Caldwell CR. Purification of mitochondrial glutamate dehydrogenase from dark-grown soybean seedlings. Plant Physiol 1996; 112:1357 - 64; PMID: 12226451
- Loulakakis KA, Roubelakis-Angelakis KA. The seven NAD(H)-glutamate dehydrogenase isoenzymes exhibit similar anabolic and catabolic activities. Physiol Plant 1996; 96:29 - 35; http://dx.doi.org/10.1111/j.1399-3054.1996.tb00179.x
- Loulakakis KA, Roubelakis-Angelakis KA. Ammonium-induced increase in NADH-glutamate dehydrogenase activity is caused by de-novo synthesis of the α-subunit. Planta 1992; 187:322 - 7; http://dx.doi.org/10.1007/BF00195655
- Turano FJ, Thakkar SS, Fang T, Weisemann JM. Characterization and expression of NAD(H)-dependent glutamate dehydrogenase genes in Arabidopsis. Plant Physiol 1997; 113:1329 - 41; http://dx.doi.org/10.1104/pp.113.4.1329; PMID: 9112779
- Fontaine JX, Saladino F, Agrimonti C, Bedu M, Tercé-Laforgue T, Tétu T, et al. Control of the synthesis and subcellular targeting of the two GDH genes products in leaves and stems of Nicotiana plumbaginifolia and Arabidopsis thaliana. Plant Cell Physiol 2006; 47:410 - 8; http://dx.doi.org/10.1093/pcp/pcj008; PMID: 16418233
- Shaw CR. The use of genetic variation in the analysis of isozyme structure. Brookhaven Symp Biol 1964; 17:117 - 30; PMID: 14246256
- Leech RM, Kirk PR. An NADP-dependent L-glutamate dehydrogenase from chloroplasts of Vicia faba L. Biochem Biophys Res Commun 1968; 32:685 - 90; http://dx.doi.org/10.1016/0006-291X(68)90293-3; PMID: 4386656
- Yakovleva VI, Kretovich VL. Gil’Manov MK. Localization of glutamate dehydrogenase in corn roots. Biokhimiya 1964; 29:401 - 6
- Lea PJ, Thurman DA. Intracellular location and properties of plant L-glutamate dehydrogenases. J Exp Bot 1972; 23:440 - 9; http://dx.doi.org/10.1093/jxb/23.2.440
- Grabowska A, Nowicki M, Kwinta J. Glutamate dehydrogenase of the germinating triticale seeds: gene expression, activity distribution and kinetic characteristics. Acta Physiol Plant 2011; 33:1981 - 90; http://dx.doi.org/10.1007/s11738-011-0801-1
- Cammaerts D, Jacobs M. A study of the role of glutamate dehydrogenase in the nitrogen metabolism of Arabidopsis thaliana.. Planta 1985; 163:517 - 26; http://dx.doi.org/10.1007/BF00392709
- Miyashita Y, Good AG. NAD(H)-dependent glutamate dehydrogenase is essential for the survival of Arabidopsis thaliana during dark-induced carbon starvation. J Exp Bot 2008; 59:667 - 80; http://dx.doi.org/10.1093/jxb/erm340; PMID: 18296429
- Yamada K, Lim J, Dale JM, Chen HM, Shinn P, Palm CJ, et al. Empirical analysis of transcriptional activity in the Arabidopsis genome. Science 2003; 302:842 - 6; http://dx.doi.org/10.1126/science.1088305; PMID: 14593172
- Igarashi D, Izumi Y, Dokiya Y, Totsuka K, Fukusaki E, Ohsumi C. Reproductive organs regulate leaf nitrogen metabolism mediated by cytokinin signal. Planta 2009; 229:633 - 44; http://dx.doi.org/10.1007/s00425-008-0858-2; PMID: 19048287
- Qiu X, Xie W, Lian X, Zhang Q. Molecular analyses of the rice glutamate dehydrogenase gene family and their response to nitrogen and phosphorous deprivation. Plant Cell Rep 2009; 28:1115 - 26; http://dx.doi.org/10.1007/s00299-009-0709-z; PMID: 19430792