Abstract
We recently demonstrated that cell elongation in plants is regulated by a triantagonistic bHLH system, in which three bHLH proteins, Activator of Cell Elongation 1 (ACE1), Arabidopsis ILI1 binding BHLH 1 (AtIBH1) and Paclobutrazol Resistance 1 (PRE1), competitively regulate the expression of genes for cell elongation. Here we show that ATBS1 Interacting Factor 2 (AIF2), AIF3 and AIF4 interact with PRE1 and ACE1, similar to AtIBH1, and also negatively regulate cell elongation in the triantagonistic bHLH system. The expression of each AIF is constitutive or induced by light, but AtIBH1 expression is dependent on BR signaling and developmental phase. These results indicate that AIFs and AtIBH1 may play different roles in cell elongation in different signaling pathways.
In plants, multiple phytohormones, namely brassinosteroids (BR), gibberellins (GA) and auxin, positively regulate cell elongation throughout the plant life cycle.Citation1 Three atypical bHLH proteins of the PRE sub-family, Paclobutrazol Resistance 1 (PRE1), PRE3/Activation-Tagged BRI1 Suppressor 1 (PRE3/ATBS1) and PRE6/KIDARI, positively regulate organ elongation in response to GA, BR and light signaling, respectively.Citation2-Citation4 These PRE sub-family proteins interact with other bHLH proteins, including Arabidopsis ILI1 Binding bHLH 1 (AtIBH1), ATBS1 Interacting Factors (AIFs) and Long Hypocotyl in Far-Red (HFR1), which negatively regulate organ elongation.Citation2,Citation4,Citation5 We recently showed that AtIBH1, PRE1 and newly identified bHLH transcriptional factors, named Activators for Cell Elongation 1 (ACE1), ACE2, ACE3 and CIB5, constitute a triantagonistic bHLH system. In this system, AtIBH1, PRE1 and ACEs competitively regulate cell elongation under BR, GA and developmental phase dependent signals.Citation6 ACE1 directly activates the expression of enzyme genes for cell elongation by interacting with their promoter regions, while AtIBH1 interacts with ACEs, interfering with the binding of ACEs to the promoters of their target genes, and thereby inhibiting cell elongation. PRE1 interacts with AtIBH1 and counteracts the ability of AtIBH1 to affect ACEs. Therefore, PRE1 restores the transcriptional activity of ACE1, resulting in induction of cell elongation.Citation6
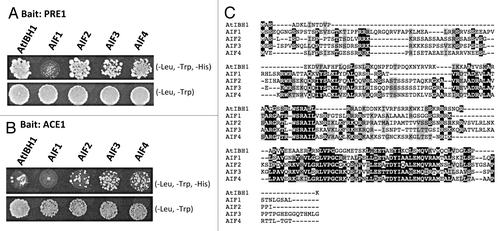
To identify other negative regulators that interact with PRE1 and function like AtIBH1 in the triantagonistic system, we performed a yeast two-hybrid (Y2H) screen using PRE1 as a bait and a library composed of only cDNAs of Arabidopsis transcription factor genes.Citation7 From this screen, we found that AIF2, AIF3 and AIF4, but not AIF1, interact with PRE1 (). We confirmed the interaction of PRE1 with AIF2, AIF3 and AIF4 by individual Y2H assays () and found that those AIFs also interacted with ACE1 (). Interestingly, AIF1, which is closely related to AIF2/3/4, does not interact with either PRE1 or ACE1 in our Y2H assays (). The AIFs are closely related to AtIBH1 and were shown to interact with PRE3/ATBS1 ().Citation3,Citation4 Wang et al. showed that AIF1 regulates cell elongation negatively but PRE3/ATBS1, which interacts with AIF1, regulates it positively in response to BR signaling.Citation4 Overexpression of AIF1 was shown to induce a dwarf phenotype, similar to the phenotype of AtIBH1-overexpressing plants.Citation4-Citation6
Table 1. TFs that interact with PRE1, as identified by yeast two-hybrid screening
Figure 1. Interaction between PRE1 and AIFs and between ACE1 and AIFs in yeast two-hybrid assays on -L-W-H and -L-W media using PRE1 (A) or ACE1 (B) as bait. (C) Alignment of amino acid sequences of AtIBH1 and each AIF.
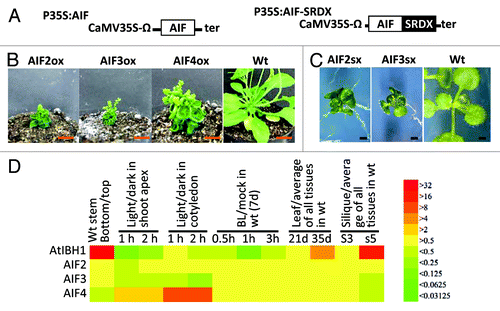
We prepared transgenic plants that ectopically expressed AIF2, AIF3 and AIF4 (P35S:AIF2, P35S:AIF3, P35S:AIF4), respectively (). All the AIF overexpressing plants exhibited drastic dwarfism, and had round-shaped, dark green leaves and short petioles and siliques (). These phenotypes were similar to the phenotype of P35S:AtIBH1 plants,Citation5,Citation6 suggesting that AIF2, AIF3 and AIF4 have similar biological functions to AtIBH1, which negatively regulates cell elongation. In addition, we analyzed the phenotype of transgenic plants that express the chimeric repressors for AIFs, which have the plant-specific SRDX repression domain fused to AIF2 and AIF3 (P35S:AIF2-SRDX and P35S:AIF3-SRDX; ).Citation8 We found that both P35S:AIF2-SRDX and P35S:AIF3-SRDX had round-shaped and dark green leaves and exhibited similar phenotypes to P35S:AIF2 and P35S:AIF3 plants (). These results indicate that these AIFs act as transcriptional repressors, similar to AtIBH1, because fusion of the SRDX to a native repressor results in a similar phenotype to its ectopic expression.Citation9
Figure 2. Functional analysis of AIF2, AIF3 and AIF4. (A) Schematic representation of the constructs. (B) Rosette plants of P35S:AIF2 (AIF2ox), P35S:AIF3 (AIF3ox), P35S:AIF4 (AIF4ox) and wild-type (wt) plants. Bar = 1 cm. (C) Fourteen-day-old seedlings of P35S:AIF2-SRDX (AIF2sx) and P35S:AIF3-SRDX (AIF3sx). Bar = 1 mm. (D) The fold change of expression of AtIBH1 and AIFs in different tissues (the bottom / top part of stem, young and old leaves and young and old siliques shoot), different light conditions in two tissues (shoot apex and cotyledon) and treatment with brassinolide (BL). Color represents the fold-change of expression to control as shown in right.
The basic motif of AIF proteins lacks the amino acids necessary for binding to the E- and G-box; therefore, it has been suggested that AIFs might not be DNA-binding bHLH proteins.Citation4,Citation10 Our results suggest that the functional mechanisms of AIF2, AIF3 and AIF4 are likely to be the same as that of AtIBH1, binding to another bHLH protein to interfere with its function. AIFs likely interfere with the DNA binding of ACE1 and inhibit cell elongation by suppressing the activation activity of ACE1. Moreover, we showed that AIFs interact with PRE; therefore, similar to its inhibition of AtIBH1, PRE is also likely to inhibit AIFs.
In this study, we show that Arabidopsis AIF2, AIF3 and AIF4 function as negative regulators of cell elongation and are likely to act redundantly with AtIBH1. AIFs may constitute a triantagonistic bHLH system with ACE1 and PRE1. AIF2/3/4, however, may regulate cell elongation differentially from AtIBH1, because the AIFs and AtIBH1 have different expression patterns ().Citation5,Citation6 For example, AIF4 expression was induced by light treatment but AIF2 and AIF3 expression was not, suggesting that AIF4 may regulate cell elongation in response to light signaling (). Also, BR and developmental phase did not affect AIF expression. These AIF expression patterns indicated that AtIBH1 and AIFs might have different mechanisms regulating their expression. Therefore AtIBH1 and each AIF may negatively regulate cell elongation under different signaling pathways, but may act through similar functional mechanisms, suppressing cell elongation via the inhibition of ACE1 DNA binding. Further studies of the systems that control the expression and post-transcriptional regulation of AIFs will provide more information on plant cell elongation.
Materials and Methods
Construction of plasmids
For construction of P35S:AIF2, cDNAs of AIF2, with the stop codon, were amplified by a pair of primers with an attB1/B2 site (attB1_At3g06590; 5′- GGGGACAAGTTTGTACAAAAAAGCAGGCTCCATGGCGTCTCTGATCTCAGATATTGAACC-3′, attB2_At3g06590; 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCTCAAATCGGTGGAGGAGCTGAGCCGTAGGA-3′) and cloned into the pDONR207 vector (Life Technologies Inc.) by BP cloning. Cloned cDNAs were transferred into pDEST_35S_HSP_GWB5 by LR clonase. For construction of P35S:AIF3 and P35S:AIF4, cDNAs of AIF3 and AIF4 with the stop codon were amplified by a pair of primers (At3g17100N; 5′- gATGGAGTCTATATCTCCGGTATCGAATCA -3′, At3g17100.2SC; 5′- GGGGTCGACTAACCAAGCATGTGTGTTTGTCCCCCTTC-3′, At1g09250N; 5′- GATGGTGGAGTCTCTGTTCCCGAGCATCGA-3′, AIF4oxsal; 5′- GGGTCGACTTAAGTTCCGGTCAACGTCGTCCGTGGTGC -3′) and cloned into the P35S vector. Constructions for CRES-T, cDNAs of AIF2 and AIF3 without the stop codon were amplified by a pair of primers (At3g06590N; 5′- gATGGCGTCTCTGATCTCAGATATTGAACC-3′, At3g06590C; 5′ AATCGGTGGAGGAGCTGAGCCGTAGGAGGA-3′, At3g17100N; 5′- gATGGAGTCTATATCTCCGGTATCGAATCA -3′, At3g17100C; 5′- ACCAAGCATGTGTGTTTGTCCCCCTTCGTG-3′) and cloned into the SmaI site of the p35SSRDXG vector.Citation11 Cloned cDNAs were transferred into pBCKH by LR clonase.
Yeast two-hybrid screening
The yeast two-hybrid assays were performed as described previously.Citation7 For bait construction, PRE1 cDNA with the stop codon was amplified by a pair of primers with an attB1/B2 site (attB1_AT5G39860.1; 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTCCATGTCGAACAGAAGATCAAGGCAATCTTC-3′, attB2_AT5G39860.1; 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCTTACATGAGTAGGCTTCTAATAACGGCGGC-3′) and ACE1 cDNA with the stop codon was amplified by a pair of primers with an attB1/B2 site (attB1_ACE1; 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTCCATGGATTTAAGTGCGAAAGATGAGTTTTCA-3′, attB2_ACE1; 5′- GGGGACCACTTTGTACAAGAAAGCTGGGTCTGGCTCAACCTTCATATTTGCAGCTGGAAG-3′) and cloned into the pDONR207 vector (Life Technologies Inc., USA) by BP cloning. Cloned cDNAs were transferred into pDEST_BTM116 by LR clonase.
Acknowledgments
The authors thank Ms. Yoko Ooi, Ms. Fumie Tobe and Ms. Miyoko Yamada for their skilled technical assistance, and Ms. Yoshimi Sugimoto and Ms. Yukie Kimura (National Institute of Advanced Industrial Science and Technology) for cultivation of plants. This work was partially supported by Grant-in-Aid for JSPS Restart Postdoctoral Fellowships.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Depuydt S, Hardtke CS. Hormone signalling crosstalk in plant growth regulation. Curr Biol 2011; 21:R365 - 73; http://dx.doi.org/10.1016/j.cub.2011.03.013; PMID: 21549959
- Hyun Y, Lee I. KIDARI, encoding a non-DNA Binding bHLH protein, represses light signal transduction in Arabidopsis thaliana.. Plant Mol Biol 2006; 61:283 - 96; http://dx.doi.org/10.1007/s11103-006-0010-2; PMID: 16786307
- Lee S, Lee S, Yang KY, Kim YM, Park SY, Kim SY, et al. Overexpression of PRE1 and its homologous genes activates Gibberellin-dependent responses in Arabidopsis thaliana.. Plant Cell Physiol 2006; 47:591 - 600; http://dx.doi.org/10.1093/pcp/pcj026; PMID: 16527868
- Wang H, Zhu Y, Fujioka S, Asami T, Li J, Li J. Regulation of Arabidopsis brassinosteroid signaling by atypical basic helix-loop-helix proteins. Plant Cell 2009; 21:3781 - 91; http://dx.doi.org/10.1105/tpc.109.072504; PMID: 20023194
- Zhang LY, Bai MY, Wu J, Zhu JY, Wang H, Zhang Z, et al. Antagonistic HLH/bHLH transcription factors mediate brassinosteroid regulation of cell elongation and plant development in rice and Arabidopsis. Plant Cell 2009; 21:3767 - 80; http://dx.doi.org/10.1105/tpc.109.070441; PMID: 20009022
- Ikeda M, Fujiwara S, Mitsuda N, Ohme-Takagi M. A triantagonistic basic helix-loop-helix system regulates cell elongation in Arabidopsis. Plant Cell 2012; 24:4483 - 97; http://dx.doi.org/10.1105/tpc.112.105023; PMID: 23161888
- Mitsuda N, Ikeda M, Takada S, Takiguchi Y, Kondou Y, Yoshizumi T, et al. Efficient yeast one-/two-hybrid screening using a library composed only of transcription factors in Arabidopsis thaliana.. Plant Cell Physiol 2010; 51:2145 - 51; http://dx.doi.org/10.1093/pcp/pcq161; PMID: 20980269
- Hiratsu K, Matsui K, Koyama T, Ohme-Takagi M. Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J 2003; 34:733 - 9; http://dx.doi.org/10.1046/j.1365-313X.2003.01759.x; PMID: 12787253
- Ikeda M, Ohme-Takagi M. A novel group of transcriptional repressors in Arabidopsis. Plant Cell Physiol 2009; 50:970 - 5; http://dx.doi.org/10.1093/pcp/pcp048; PMID: 19324928
- Toledo-Ortiz G, Huq E, Quail PH. The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 2003; 15:1749 - 70; http://dx.doi.org/10.1105/tpc.013839; PMID: 12897250
- Mitsuda N, Hiratsu K, Todaka D, Nakashima K, Yamaguchi-Shinozaki K, Ohme-Takagi M. Efficient production of male and female sterile plants by expression of a chimeric repressor in Arabidopsis and rice. Plant Biotechnol J 2006; 4:325 - 32; http://dx.doi.org/10.1111/j.1467-7652.2006.00184.x; PMID: 17147638