Abstract
The translocation of effector proteins into the host plant cells is essential for pathogens to suppress plant immune responses. The oomycete pathogen Phytophthora infestans secretes AVR3a, a crucial virulence effector protein with an N-terminal RXLR motif that is required for this translocation. It has been reported that the RXLR motif of P. sojae Avr1b, which is a close homolog of AVR3a, is required for binding to phosphatidylinositol monophosphates (PIPs). However, in our previous report, AVR3a as well as Avr1b bind to PIPs not via RXLR but via lysine residues forming a positively-charged area in the effector domain. In this report, we examined whether other RXLR effectors whose structures have been determined bind to PIPs. Both P. capsici AVR3a11 and Hyaloperonospora arabidopsidis ATR1 have an RXLR motif in their N-terminal regions but did not bind to any PIPs. These results suggest that the RXLR motif is not sufficient for PIP binding.
Filamentous plant pathogens, including oomycete and fungi, secrete a number of effector proteins that accumulate in apoplastic spaces or enter host plant cells to modulate host immune responses.Citation1,Citation2 AVR3a, an effector protein secreted from the oomycete pathogen Phytophthora infestans causing potato late blight disease, has the characteristic RXLR motif sequence of amino acids Arg-X-Leu-Arg (where X is any amino acid) at the N-terminus and an effector domain harboring virulence functions at the C-terminal end. AVR3a is translocated into the host cells in an RXLR-motif dependent manner.Citation3,Citation4
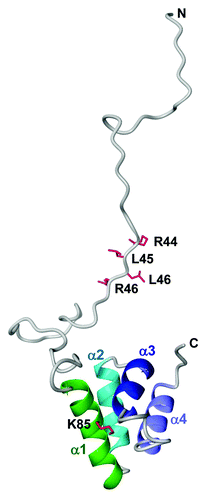
In our previous work, we determined the protein structure of P. capsici AVR3a4, which is a close homolog of AVR3a, by NMR analysis.Citation5 The NMR-derived model structure of AVR3a showed that the effector domain comprises four α-helices, but the N-terminal region including the RXLR motif is disordered (). Kale et al.Citation6 have reported that the RXLR motif of Avr1b, which is a close homolog of AVR3a in P. sojae, binds to phosphatidylinositol monophosphate (PIPs) lipids on the surface of host cells and hypothesized that this binding is required for pathogen-independent entry of the protein into host cells. On the contrary, we found that AVR3a as well as Avr1b bound to PIPs not via the RXLR motif, but via lysine residues forming a positively-charged area in the effector domain.Citation5 In agreement with our findings, it was recently shown that the PIP-binding abilities of AVR3a are mediated by its effector domain, not its RXLR motif.Citation7 However, it was reported that the MiSSP7 effector protein secreted from Laccaria bicolor, a mutualistic ectomycorrhizal symbiont of poplar, can enter host plant cells via an RXLR-like motif, RALG, and that the motif binds to PIPs.Citation8 To resolve these discrepancies, it is necessary to perform further studies on whether other RXLR effectors also bind to PIPs.
Figure 1. The NMR-derived model structure of AVR3a with the RXLR motif. The residues for the RXLR motif (R44, L45, L46 and R47) and PIP binding (K85) are mapped on the ribbon diagram as red sticks.
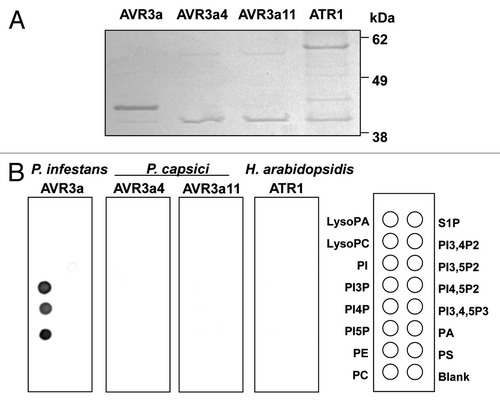
In addition to AVR3a4, the protein structures of AVR3a11 from P. capsici as well as ATR1 from Hyaloperonospora arabidopsidis, an oomycete downy mildew pathogen, were determined.Citation9,Citation10 We therefore investigated whether these RXLR effectors bind to PIPs. The GST fusions of these proteins were used for a lipid overlay assay as described in Yaeno et al.Citation5 As shown in , AVR3a11 and ATR1 does not bind to PIPs, even though they harbor the RXLR motif at the N-terminus, suggesting that the RXLR motif is insufficient for PIP binding. These RXLR effectors also have the WY motif in the effector domain as a conserved structural fold.Citation11 Thus, the WY motif is unlikely to be involved in PIP binding.
Figure 2. Lipid overlay assay of oomycete RXLR effectors, AVR3a, AVR3a4, AVR3a11 and ATR1. (A) Escherichia coli strain BL21-AI was transformed with pDEST24 constructs for AVR3a (Asp23-Tyr147), AVR3a4 (Asn22-Tyr122), AVR3a11 (Asn22-Val132) or ATR1-Emwa1 (Ser22-Glu324). Protein expression and purification were performed as described in Yaeno et al.Citation5 The purified C-terminal GST fusion proteins were checked by SDS-PAGE stained with InstantBlue (Expedeon) and equal amounts of proteins were used for the lipid overlay assay. (B) Nitrocellulose membranes spotted with 100 pmol of various lipids (PIP Strips; Echelon Biosciences) were blocked in 1% nonfat milk in PBS for 1 h and then incubated with 1 μg/mL C-terminal GST fusions of P. infestans AVR3a, P. capsici AVR3a4, P. capsici AVR3a11 and H. arabidopsidis ATR1 overnight at 4°C. After washing with PBS-T, the bound proteins were detected using anti–GST-HRP antibodies (GE Healthcare) diluted to 1:2,000. PA, phosphatidic acid; PC, phosphatidyl-choline; PE, phosphatidylethanolamine; PI, phosphatidylinositol; PI3P, PI-3-phosphate; PI4P, PI-4-phosphate; PI5P, PI-5-phosphate; PI3,4P2, PI-3,4-biphosphate; PI3,5P2, PI-3,5-biphosphate; PI4,5P2, PI-4,5-biphosphate; PI3,4,5P3, PI-3,4,5-triphosphate; PS, phosphatidylserine; S1P, sphingosine-1-phosphate.
Recently, consistent in principle with our findings,Citation5 Sun et al.Citation12 showed that the P. sojae RXLR effector Avh5 bound to PIPs predominantly via the lysine residues of the C-terminal effector domain. The mutations in the RXLR motif of Avh5 did not have much effect on PIP binding. This is inconsistent with the finding by Kale et al.Citation6 showing that Avh5 binds to PIPs via the RXLR motif and the reason for this discrepancy is unclear.
The PIP-binding ability of the effector domain in AVR3a may be essential for protein stability inside the host cells.Citation5 Similarly, PIP binding confers thermal stability and a protective effect against trypsin proteolysis to Avh5. As the results were obtained from NMR analysis and circular dichroism spectra, the tested Avh5 protein is likely to be properly structured.Citation12 In contrast, it was reported that PIP binding was observed in only denatured AVR3a proteins in vitro and was physiologically irrelevant.Citation7 Thus the physiological roles of PIP binding in the effector domains remain to be elucidated.
A major point of contention in studies on effector translocation is whether or not the PIP-binding abilities of effectors are involved in host cell entry. AvrM, an effector from Melampsora lini, the flax rust fungus, enters host plant cells but has no obvious motif in the region required for entry.Citation13 Gan et al.Citation14 found that although AvrM bound to PIPs, the binding was independent of the region required for host cell entry. SpHtp1, an effector from the fish pathogenic oomycete Saprolegnia parasitica, enters fish cells in an RXLR-like motif dependent manner. However, this process is not mediated by PIP binding.Citation15 The effectors of the human malaria parasite Plasmodium falciparum which are delivered into host cells also have an RXLR-like motif RxLxE/D/Q required for translocation across the parasitiphorous vacuolar membrane into the host erythrocyte cytoplasm.Citation16,Citation17 Interestingly, the RxLxE/D/Q motif binds to PI3P in parasite endoplasmic reticulum (ER) membranes in the process of export to the erythrocyte.Citation18 However, in fact, the motif is cleaved by a protease in the parasite ER before export, and furthermore, the cell surfaces of host erythrocytes do not have detectable levels of PI3P.Citation6,Citation19,Citation20 Thus, the entry of Plasmodium effectors cannot be explained by PIP-binding of their RXLR-like motif to host PIPs. Even if the RXLR motif itself has a PIP-binding ability similar to the RxLxE/D/Q motif, the mechanism of cell entry appears to differ between RXLR effectors and Plasmodium effectors. This is because unlike Plasmodium effectors, RXLR effectors can enter host cells without the requirement of pathogen-encoded machinery.Citation4,Citation21,Citation22 Clearly, many questions remain to be resolved to elucidate the mechanisms underlying host cell entry of effector proteins, including the relationship between PIP binding and cell entry.
Acknowledgments
We thank Pamela Gan for critical reading of the manuscript; and Kaori Takizawa and Yoko Nagai for technical supports. This work was supported by JSPS KAKENHI Grant Numbers 24228008 (to K.S.) and 24780046 (to T.Y.).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Kamoun S. A catalogue of the effector secretome of plant pathogenic oomycetes. Annu Rev Phytopathol 2006; 44:41 - 60; http://dx.doi.org/10.1146/annurev.phyto.44.070505.143436; PMID: 16448329
- Oliva R, Win J, Raffaele S, Boutemy L, Bozkurt TO, Chaparro-Garcia A, et al. Recent developments in effector biology of filamentous plant pathogens. Cell Microbiol 2010; 12:705 - 15; http://dx.doi.org/10.1111/j.1462-5822.2010.01471.x; PMID: 20374248
- Birch PRJ, Boevink PC, Gilroy EM, Hein I, Pritchard L, Whisson SC. Oomycete RXLR effectors: delivery, functional redundancy and durable disease resistance. Curr Opin Plant Biol 2008; 11:373 - 9; http://dx.doi.org/10.1016/j.pbi.2008.04.005; PMID: 18511334
- Whisson SC, Boevink PC, Moleleki L, Avrova AO, Morales JG, Gilroy EM, et al. A translocation signal for delivery of oomycete effector proteins into host plant cells. Nature 2007; 450:115 - 8; http://dx.doi.org/10.1038/nature06203; PMID: 17914356
- Yaeno T, Li H, Chaparro-Garcia A, Schornack S, Koshiba S, Watanabe S, et al. Phosphatidylinositol monophosphate-binding interface in the oomycete RXLR effector AVR3a is required for its stability in host cells to modulate plant immunity. Proc Natl Acad Sci USA 2011; 108:14682 - 7; http://dx.doi.org/10.1073/pnas.1106002108; PMID: 21821794
- Kale SD, Gu B, Capelluto DGS, Dou D, Feldman E, Rumore A, et al. External lipid PI3P mediates entry of eukaryotic pathogen effectors into plant and animal host cells. Cell 2010; 142:284 - 95; http://dx.doi.org/10.1016/j.cell.2010.06.008; PMID: 20655469
- Wawra S, Agacan M, Boddey JA, Davidson I, Gachon CMM, Zanda M, et al. Avirulence protein 3a (AVR3a) from the potato pathogen Phytophthora infestans forms homodimers through its predicted translocation region and does not specifically bind phospholipids. J Biol Chem 2012; 287:38101 - 9; http://dx.doi.org/10.1074/jbc.M112.395129; PMID: 22977236
- Plett JM, Kemppainen M, Kale SD, Kohler A, Legué V, Brun A, et al. A secreted effector protein of Laccaria bicolor is required for symbiosis development. Curr Biol 2011; 21:1197 - 203; http://dx.doi.org/10.1016/j.cub.2011.05.033; PMID: 21757352
- Boutemy LS, King SRF, Win J, Hughes RK, Clarke TA, Blumenschein TMA, et al. Structures of Phytophthora RXLR effector proteins: a conserved but adaptable fold underpins functional diversity. J Biol Chem 2011; 286:35834 - 42; http://dx.doi.org/10.1074/jbc.M111.262303; PMID: 21813644
- Chou S, Krasileva KV, Holton JM, Steinbrenner AD, Alber T, Staskawicz BJ. Hyaloperonospora arabidopsidis ATR1 effector is a repeat protein with distributed recognition surfaces. Proc Natl Acad Sci USA 2011; 108:13323 - 8; http://dx.doi.org/10.1073/pnas.1109791108; PMID: 21788488
- Win J, Krasileva KV, Kamoun S, Shirasu K, Staskawicz BJ, Banfield MJ. Sequence divergent RXLR effectors share a structural fold conserved across plant pathogenic oomycete species. PLoS Pathog 2012; 8:e1002400; http://dx.doi.org/10.1371/journal.ppat.1002400; PMID: 22253591
- Sun F, Kale SD, Azurmendi HF, Li D, Tyler BM, Capelluto DGS. Structural basis for interactions of the Phytophthora sojae RxLR effector Avh5 with phosphatidylinositol 3-phosphate and for host cell entry. Mol Plant Microbe Interact 2012; In press http://dx.doi.org/10.1094/MPMI-07-12-0184-R; PMID: 23075041
- Rafiqi M, Gan PHP, Ravensdale M, Lawrence GJ, Ellis JG, Jones DA, et al. Internalization of flax rust avirulence proteins into flax and tobacco cells can occur in the absence of the pathogen. Plant Cell 2010; 22:2017 - 32; http://dx.doi.org/10.1105/tpc.109.072983; PMID: 20525849
- Gan PHP, Rafiqi M, Ellis JG, Jones DA, Hardham AR, Dodds PN. Lipid binding activities of flax rust AvrM and AvrL567 effectors. Plant Signal Behav 2010; 5:1272 - 5; http://dx.doi.org/10.4161/psb.5.10.13013; PMID: 20855950
- Wawra S, Bain J, Durward E, de Bruijn I, Minor KL, Matena A, et al. Host-targeting protein 1 (SpHtp1) from the oomycete Saprolegnia parasitica translocates specifically into fish cells in a tyrosine-O-sulphate-dependent manner. Proc Natl Acad Sci USA 2012; 109:2096 - 101; http://dx.doi.org/10.1073/pnas.1113775109; PMID: 22308362
- Hiller NL, Bhattacharjee S, van Ooij C, Liolios K, Harrison T, Lopez-Estraño C, et al. A host-targeting signal in virulence proteins reveals a secretome in malarial infection. Science 2004; 306:1934 - 7; http://dx.doi.org/10.1126/science.1102737; PMID: 15591203
- Marti M, Good RT, Rug M, Knuepfer E, Cowman AF. Targeting malaria virulence and remodeling proteins to the host erythrocyte. Science 2004; 306:1930 - 3; http://dx.doi.org/10.1126/science.1102452; PMID: 15591202
- Bhattacharjee S, Stahelin RV, Speicher KD, Speicher DW, Haldar K. Endoplasmic reticulum PI(3)P lipid binding targets malaria proteins to the host cell. Cell 2012; 148:201 - 12; http://dx.doi.org/10.1016/j.cell.2011.10.051; PMID: 22265412
- Boddey JA, Hodder AN, Günther S, Gilson PR, Patsiouras H, Kapp EA, et al. An aspartyl protease directs malaria effector proteins to the host cell. Nature 2010; 463:627 - 31; http://dx.doi.org/10.1038/nature08728; PMID: 20130643
- Russo I, Babbitt S, Muralidharan V, Butler T, Oksman A, Goldberg DE. Plasmepsin V licenses Plasmodium proteins for export into the host erythrocyte. Nature 2010; 463:632 - 6; http://dx.doi.org/10.1038/nature08726; PMID: 20130644
- de Koning-Ward TF, Gilson PR, Boddey JA, Rug M, Smith BJ, Papenfuss AT, et al. A newly discovered protein export machine in malaria parasites. Nature 2009; 459:945 - 9; http://dx.doi.org/10.1038/nature08104; PMID: 19536257
- Dou D, Kale SD, Wang X, Jiang RHY, Bruce NA, Arredondo FD, et al. RXLR-mediated entry of Phytophthora sojae effector Avr1b into soybean cells does not require pathogen-encoded machinery. Plant Cell 2008; 20:1930 - 47; http://dx.doi.org/10.1105/tpc.107.056093; PMID: 18621946