Abstract
NB-LRR-type disease resistance (R) genes have been used in traditional breeding programs for crop protection. However, functional transfer of NB-LRR-type R genes to plants in taxonomically distinct families to establish pathogen resistance has not been successful. Here we demonstrate that a pair of Arabidopsis (Brassicaceae) NB-LRR-type R genes, RPS4 and RRS1, properly function in two other Brassicaceae, Brassica rapa and B. napus, but also in two Solanaceae, Nicotiana benthamiana and tomato (Solanum lycopersicum). The solanaceous plants transformed with RPS4/RRS1 confer bacterial effector-specific immunity responses. Furthermore, RPS4 and RRS1, which confer resistance to a fungal pathogen Colletotrichum higginsianum in Brassicaceae, also protect against Colletotrichum orbiculare in cucumber (Cucurbitaceae). Thus the successful transfer of two R genes at the family level overcomes restricted taxonomic functionality. This implies that the downstream components of R genes must be highly conserved and interfamily utilization of R genes can be a powerful strategy to combat pathogens.
Plants trigger innate immunity responses to pathogens via a two-layer surveillance system composed of pattern recognition receptors (PRRs) and nucleotide binding-leucine rich repeat (NB-LRR) proteins that are encoded by resistance (R) genes.Citation1 PRRs recognize microbe-associated molecular patterns (MAMPs) at a plasma membrane, and NB-LRR proteins subsequently detect pathogen-derived effectors inside the cell. Although interfamily transfer of PRR-mediated disease resistance has been successful,Citation2 no R genes have been successfully expressed in a different family, a phenomenon which has come to be known as restricted taxonomic functionality (RTF) of R genes.Citation3 Heterologous expression of NB-LRR type R genes in a taxonomically distinct family triggers either no response or inappropriate auto-immunity responses, suggesting that the regulatory or signaling components associated with NB-LRR protein-based resistance are family specific.Citation4
A pair of Arabidopsis thaliana (Brasicaceae) NB-LRR type R genes, RPS4 and RRS1, function together to confer disease resistance against two taxonomically distinct bacteria, Pseudomonas syringae pv tomato DC3000, which produces the effector AvrRps4 (Pst-avrRps4), and Ralstonia solanacearum strains, which express the PopP2 effectorCitation5 (). To determine whether the RPS4/RRS1 R gene pair also functions in non-Brasicaceae plants, we generated transgenic Nicotiana benthamiana (Solanaceae) plants expressing RPS4 and RRS1 under control of their cognate promoters. We found that either of the two bacterial effectors, AvrRps4 or PopP2 produced in planta via Agrobacterium-mediated transient expression, induced cell death in N. benthamiana transformed with both R genes (RPS4+RRS1), but not in plants expressing only RPS4 or RRS1. Importantly, the transgenic N. benthamiana plants showed no significant constitutive expression of inducible defense-related genes, indicating that the conferred resistance is effector specific.
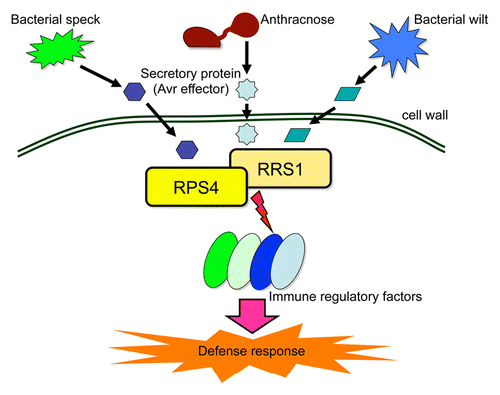
Figure 1.RPS4 and RRS1 function as a dual resistance gene system that prevents infection by three distinct pathogens (Anthracnose; Colletotrichum higginsianum, Bacterial wilt; Ralstonia solanacearum, and Bacterial speck; Pseudomonas syringae pv tomato strain DC3000 expressing avrRps4). Dual resistance (R) proteins recognize corresponding avirulence (Avr) effectors either indirectly through detection of changes in their host protein targets or through direct dual R protein-Avr effector interaction.
R. solanacearum causes bacterial wilt, a serious soilborne disease of many plants worldwide. Resistance lines are urgently needed, as natural resource for resistance is limited and soil fumigation has not been effective. Tomato plants (Solanum lycopersicum) transformed with RPS4 and RRS1 also conferred resistance to R. solanacearum expressing popP2, indicating that the conferred resistance is specific for the PopP2 effector. Pst-avrRps4 causes bacterial speck on tomato, a disease characterized by defoliation, blossom blight and lesions on developing fruit. As in Arabidopsis, the transgenic tomato plants exhibited resistance against the Pst-avrRps4 pathogen, but showed no significant constitutive expression of inducible defense-related genes, indicating that the conferred resistance is specific for the AvrRps4 effector. Thus, RPS4 and RRS1 are functional in at least two solanaceous plants, N. benthamiana and tomato.
In Arabidopsis, the dual RPS4/RRS1 genes also confer resistance to the fungal pathogen Colletotrichum higginsianum.Citation5 Colletotrichum spp cause anthracnose disease in a wide range of host plants, including cucumber (Cucumis sativus, Cucurbitaceae). We generated transgenic cucumber plants expressing RPS4/RRS1 and inoculated them with Colletotrichum orbiculare, which infects cucurbits. WT cucumber plants developed brown necrotic lesions surrounded by a yellow halo, a typical symptom of anthracnose disease. RPS4/RRS1 plants were highly resistant, developing only small necrotic flecks at the inoculated sites, indicative of an active defense reaction. Transgenic plants grew normally and did not express inducible defense-related genes, suggesting that autoimmunity is not induced by RPS4/RRS1 in cucumber. These data indicate that RPS4/RRS1 recognize effectors common to Colletotrichum or detect some alteration of a host protein targeted by both strains.
Our study demonstrates that introduction of the two RPS4 and RRS1 overcomes RTF and suggests that the downstream components of the R genes are highly conserved. It is likely that R gene-based immunity can be transferred to distantly related species once the right gene pair is identified. The number of known potential pairs of R genes from various plant species is increasingCitation6 and we postulate that some of those pairs may also overcome RTF. In summary, this finding indicates that a new strategy can be used for creating pathogen-resistant vegetables and crops by using a previously unexploited resource of durable genetic resistance.
Acknowledgments
This work was supported by the Programme for Promotion of Basic and Applied Researches for Innovations in Bio-oriented Industry to K.S., Y.Tak., Y.Nar. and by Grant-in-Aid for Scientific Research (KAKENHI) (21580060 to Y.Nar., 21780038 to M.N. and 19678001 to K.S.). We thank Mariko Miyashita, Yoko Iwasaki and Yasuyo Katayama for their excellent technical assistance and Tsuyoshi Nakagawa (Shimane University) for kindly providing pGWB1. The tomato resources used in this research were provided by the National BioResource Project (NBRP), Japanese Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan. The production of transgenic tomato plants was supported by the RIKEN Plant Transformation Network.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Jones JDG, Dangl JL. The plant immune system. Nature 2006; 444:323 - 9; http://dx.doi.org/10.1038/nature05286; PMID: 17108957
- Lacombe S, Rougon-Cardoso A, Sherwood E, Peeters N, Dahlbeck D, van Esse HP, et al. Interfamily transfer of a plant pattern-recognition receptor confers broad-spectrum bacterial resistance. Nat Biotechnol 2010; 28:365 - 9; http://dx.doi.org/10.1038/nbt.1613; PMID: 20231819
- Tai TH, Dahlbeck D, Clark ET, Gajiwala P, Pasion R, Whalen MC, et al. Expression of the Bs2 pepper gene confers resistance to bacterial spot disease in tomato. Proc Natl Acad Sci USA 1999; 96:14153 - 8; http://dx.doi.org/10.1073/pnas.96.24.14153; PMID: 10570214
- Brutus A, He SY. Pattern recognition receptors: going practical. Nat Biotechnol 2010; 28:330 - 1; http://dx.doi.org/10.1038/nbt0410-330; PMID: 20379175
- Narusaka M, Shirasu K, Noutoshi Y, Kubo Y, Shiraishi T, Iwabuchi M, et al. RRS1 and RPS4 provide a dual Resistance-gene system against fungal and bacterial pathogens. Plant J 2009; 60:218 - 26; http://dx.doi.org/10.1111/j.1365-313X.2009.03949.x; PMID: 19519800
- Eitas TK, Dangl JL. NB-LRR proteins: pairs, pieces, perception, partners, and pathways. Curr Opin Plant Biol 2010; 13:472 - 7; http://dx.doi.org/10.1016/j.pbi.2010.04.007; PMID: 20483655