Abstract
The non-protein amino acid γ-aminobutyric acid (GABA) accumulates in plants in response to a wide variety of environmental cues. Recent data point toward an involvement of GABA in tricarboxylic acid (TCA) cycle activity and respiration, especially in stressed roots. To gain further insights into potential GABA functions in plants, phylogenetic and bioinformatic approaches were undertaken. Phylogenetic reconstruction of the GABA transaminase (GABA-T) protein family revealed the monophyletic nature of plant GABA-Ts. However, this analysis also pointed to the common origin of several plant aminotransferases families, which were found more similar to plant GABA-Ts than yeast and human GABA-Ts. A computational analysis of AtGABA-T co-expressed genes was performed in roots and in stress conditions. This second approach uncovered a strong connection between GABA metabolism and glyoxylate cycle during stress. Both in silico analyses open new perspectives and hypotheses for GABA metabolic functions in plants.
Gamma-aminobutyric acid (GABA), a four-carbon non-protein amino acid, has drawn much attention since its first identification over 60 y ago in potato tuber,Citation1 rat brainCitation2 and yeast extract.Citation3 Subsequent investigations of GABA functions followed two main directions: signaling and metabolism. Signaling effects of GABA have been intensively tackled in mammalian models. It has been clearly established that GABA acts as a neurotransmitterCitation4 and impacts the central nervous system developmentCitation5,Citation6 In plants, early studies in the 70–80s reported rapid GABA accumulation in response to many environmental cues.Citation7-Citation10 Based on these observations, it has been hypothesized that GABA could participate in stress responses and tolerance. Nevertheless, for decades, GABA functions in plants remained elusive. Major progress came along with the genomic era and availability of the powerful embedded tools. Genetic analyses in Arabidopsis demonstrated that GABA acts as a signal molecule for pollen tube guidance and elongation.Citation11,Citation12 Alteration of tricarboxylic acid (TCA) cycle enzyme activities shed light on GABA metabolic function and stressed the tight connection between GABA and respiration.Citation13-Citation15
The functional implication of GABA metabolism in Arabidopsis salt stress tolerance has recently been reported.Citation16,Citation17 The GABA transaminase gaba-t/pop2-1 mutant root growth was shown hypersensitive to the ionic component of salt stress,Citation16 confirming the long-standing hypothesis of GABA involvement in plant stress response. Metabolic data revealed that GABA bridges amino and organic acids metabolisms in roots under saline conditions.Citation16,Citation17 Moreover, a genome-wide transcriptional analysis indicated that central carbon metabolism is altered upon the loss of GABA metabolic function during salt stress. Indeed, genes involved in remobilization of carbon reserves (i.e., sucrose and starch) were found higher expressed in the gaba-t/pop2-1 mutant.Citation17 However, despite this transcriptional upregulation, a decrease in soluble sugars was observed in the mutant,Citation16,Citation17 a response that might be a compensatory effect of enhanced glycolysis balancing the reduced succinate supply due to GABA metabolism defect.Citation17 Here, GABA function was further explored by (1) investigating the phylogeny of the GABA transaminase (GABA-T) protein family, and (2) performing a computational analysis of Arabidopsis GABA-T co-expressed genes.
Phylogenetic analysis of GABA-T protein gamily
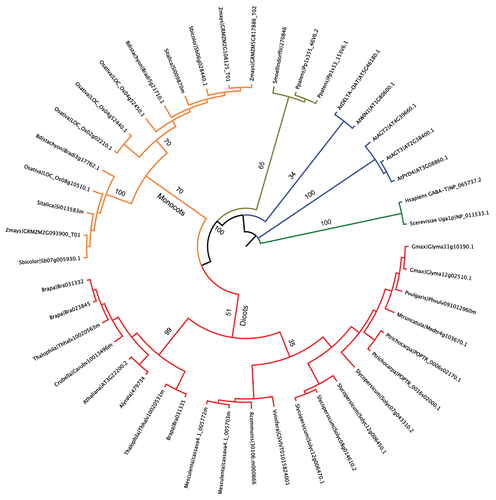
GABA-T-like protein sequences from plants, human and yeast were retrieved from Phytozome (www.phytozome.org) and GenBank (www.ncbi.nlm.nih.gov/genbank) databases based on sequence homology to Arabidopsis GABA-T (AtGABA-T; At3g22200). Protein sequences were aligned using MEGA5 software (www.megasoftware.net) and MUSCLE algorithm. Subsequently, phylogeny was reconstructed using the Neighbor-joining method (Jones-Taylor-Thornton model). Human and yeast GABA-T sequences were used to root the tree. Likewise, a set of Arabidopsis aminotransferases closely related to AtGABA-T [i.e., alanine:glyoxylate aminotransferases (AGT2 and AGT3), β-alanine aminotransferase (PYD4), ornithine delta-aminotransferase (DELTA-OAT) and acetylornithine transaminase (WIN1)] served as outgroup. The phylogenetic reconstruction clearly discriminated GABA-Ts from ancestors (i.e., Physcomitrella patens and Selaginella moellendorffii), monocot and dicot lineages. Furthermore, an early and conserved duplication of GABA-Ts was uncovered in monocots while in dicots only closely related paralogs evolved in some species (). Strikingly, the phylogenetic distance between the Arabidopsis aminotransferases and plant GABA-Ts was shorter than the distance between the functional yeast, human and plant GABA-T homologs (). This suggests that plant aminotransferases might share common substrates and activities. Arabidopsis, tomato and rice GABA-Ts, able to use both pyruvate and glyoxylate as amino group acceptor,Citation18-Citation20 exemplify the potentially broad substrate range of this class of proteins.
Figure 1. Phylogenetic relationships among plant GABA-T family. Neighbor-joining tree was constructed using the MEGA5 software. Human and yeast GABA-T protein sequences were used to root the tree. A set of Arabidopsis proteins closely related to AtGABA-T was included as outgroup. Branch lengths are proportional to phylogenetic distance; bootstrap values (expressed as percentage) from 1,000 iterations are indicated on main branches. For each protein, organism name and locus identifier/GenBank accession is provided.
Functional classification of AtGABA-T co-expressed genes
As AtGABA-T plays a prominent role in the metabolism of stressed roots,Citation16,Citation17 AtGABA-T co-expressed genes were identified in “AtGenExpress Root” and “AtGenExpress Stress” transcriptomic series using the Expression Angler software.Citation21 Applying a stringent Pearson correlation coefficient threshold of 0.8, 254 and 140 genes co-expressing with AtGABA-T were retrieved from “Root” and “Stress” data sets, respectively (lists in supplemental data). Among them, the expression of 12 genes correlated with AtGABA-T expression in both data sets (list “Root and Stress” in supplemental data). Next, a functional analysis of the three gene sets (i.e., “Root,” “Stress,” “Root and Stress”) was performed using the Classification SuperViewer softwareCitation22 and MapMan terms. Interestingly, “Polyamine metabolism,” “N metabolism” and “TCA org. transformation” terms were found more than 10-fold enriched among AtGABA-T co-expressed genes in the “Root” serie (). This result is in agreement with the previous finding that GABA links N and C metabolisms in roots.Citation16,Citation17 Other significantly enriched terms were associated with sugars (i.e., CHO), nucleotide and amino acid metabolisms, as well as with transport process and mitochondrial electron transport (). The latter observation further corroborates the previously suggested participation of GABA metabolism to root respiration.Citation17 In the gene list extracted from the “Stress” data set, “Gluconeogenesis / glyoxylate” term was almost 37-fold enriched compared with the entire genome (). Strikingly, the same term was recovered as 200 times overrepresented in the overlap between “Root” and “Stress” series (). The glyoxylate cycle allows to bypass the decarboxylation steps of TCA cycle and to use lipids as respiratory substrate or carbon source for gluconeogenesis.Citation23,Citation24 Regulation of the glyoxylate cycle genes is tuned by the carbon metabolic status of plants; the glyoxylate cycle is in particular induced under sugar starvation conditions.Citation25 Interestingly, roots of the Arabidopsis gaba-t/pop2–1 mutant are deprived in sugars after salt stress,Citation16,Citation17 a similar phenomenon is observed in plants lacking glyoxylate cycle under low light,Citation26,Citation27 two conditions known to lead to limited carbon availability. These observations raise intriguing hypotheses for GABA function in plants, especially regarding stress acclimation. The glyoxylate cycle is initiated by the isocitrate lyase that cleaves isocitrate into glyoxylate and succinate in peroxysomes. Given that AtGABA-T can use glyoxylate as amino group acceptor,Citation18 it can be hypothesized that it uses directly glyoxylate derived from isocitrate cleavage to form glycine and, after the succinic semialdehyde dehydrogenase step of GABA metabolism, a second succinate molecule. Alternatively, GABA metabolism could be embedded in a fine tuned metabolic network involving lipid and protein respiration that would allow stressed plants to keep fuelling cells with energy upon reduced carbon fixation. In support of second hypothesis, the term “Lipid metabolism” was found six times enriched among AtGABA-T co-expressed genes under stress conditions ().
Table 1. Functional classification of AtGABA-T co-expressed genes
Phylogenetic and bioinformatic data presented in this paper provide a platform to propose novel hypotheses of GABA functions in plants. In this context, it would be informative to experimentally explore the connection between GABA metabolism and glyoxylate cycle under stress conditions.
Additional material
Download Zip (1.4 MB)Acknowledgments
I am very grateful to Drs Anne Molitor and Danièle Werck-Reichhart for the critical reading of the manuscript.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplementary Material
Supplementary material may be found here: www.landesbioscience.com/journals/psb/article/24274
References
- Steward FC, Thompson JF, Dent CE. γ-Aminobutyric acid: a constituent of the potato tuber?. Science 1949; 110:439 - 40
- Roberts E, Frankel S. γ-Aminobutyric acid in brain: its formation from glutamic acid. J Biol Chem 1950; 187:55 - 63; PMID: 14794689
- Reed LJ. The occurrence of γ-aminobutyric acid in yeast extract: its isolation and identification. J Biol Chem 1950; 183:451 - 8
- Owens DF, Kriegstein AR. Is there more to GABA than synaptic inhibition?. Nat Rev Neurosci 2002; 3:715 - 27; http://dx.doi.org/10.1038/nrn919; PMID: 12209120
- Ganguly K, Schinder AF, Wong ST, Poo M-M. GABA itself promotes the developmental switch of neuronal GABAergic responses from excitation to inhibition. Cell 2001; 105:521 - 32; http://dx.doi.org/10.1016/S0092-8674(01)00341-5; PMID: 11371348
- Ge S, Goh ELK, Sailor KA, Kitabatake Y, Ming GL, Song H. GABA regulates synaptic integration of newly generated neurons in the adult brain. Nature 2006; 439:589 - 93; http://dx.doi.org/10.1038/nature04404; PMID: 16341203
- Streeter JG, Thompson JF. Anaerobic accumulation of γ-aminobutyric acid and alanine in radish leaves (Raphanus sativus, L.). Plant Physiol 1972; 49:572 - 8; http://dx.doi.org/10.1104/pp.49.4.572; PMID: 16658004
- Handa S, Bressan RA, Handa AK, Carpita NC, Hasegawa PM. Solutes contributing to osmotic adjustment in cultured plant cells adapted to water stress. Plant Physiol 1983; 73:834 - 43; http://dx.doi.org/10.1104/pp.73.3.834; PMID: 16663309
- Binzel ML, Hasegawa PM, Rhodes D, Handa S, Handa AK, Bressan RA. Solute accumulation in tobacco cells adapted to NaCl. Plant Physiol 1987; 84:1408 - 15; http://dx.doi.org/10.1104/pp.84.4.1408; PMID: 16665618
- Wallace W, Secor J, Schrader LE. Rapid accumulation of γ-aminobutyric acid and alanine in soybean leaves in response to an abrupt transfer to lower temperature, darkness, or mechanical manipulation. Plant Physiol 1984; 75:170 - 5; http://dx.doi.org/10.1104/pp.75.1.170; PMID: 16663565
- Palanivelu R, Brass L, Edlund AF, Preuss D. Pollen tube growth and guidance is regulated by POP2, an Arabidopsis gene that controls GABA levels. Cell 2003; 114:47 - 59; http://dx.doi.org/10.1016/S0092-8674(03)00479-3; PMID: 12859897
- Renault H, El Amrani A, Palanivelu R, Updegraff EP, Yu A, Renou J-P, et al. GABA accumulation causes cell elongation defects and a decrease in expression of genes encoding secreted and cell wall-related proteins in Arabidopsis thaliana.. Plant Cell Physiol 2011; 52:894 - 908; http://dx.doi.org/10.1093/pcp/pcr041; PMID: 21471118
- Araújo WL, Tohge T, Osorio S, Lohse M, Balbo I, Krahnert I, et al. Antisense inhibition of the 2-oxoglutarate dehydrogenase complex in tomato demonstrates its importance for plant respiration and during leaf senescence and fruit maturation. Plant Cell 2012; 24:2328 - 51; http://dx.doi.org/10.1105/tpc.112.099002; PMID: 22751214
- Studart-Guimarães C, Fait A, Nunes-Nesi A, Carrari F, Usadel B, Fernie AR. Reduced expression of succinyl-coenzyme A ligase can be compensated for by up-regulation of the γ-aminobutyrate shunt in illuminated tomato leaves. Plant Physiol 2007; 145:626 - 39; http://dx.doi.org/10.1104/pp.107.103101; PMID: 17885090
- Nunes-Nesi A, Carrari F, Lytovchenko A, Smith AMO, Loureiro ME, Ratcliffe RG, et al. Enhanced photosynthetic performance and growth as a consequence of decreasing mitochondrial malate dehydrogenase activity in transgenic tomato plants. Plant Physiol 2005; 137:611 - 22; http://dx.doi.org/10.1104/pp.104.055566; PMID: 15665243
- Renault H, Roussel V, El Amrani A, Arzel M, Renault D, Bouchereau A, et al. The Arabidopsis pop2-1 mutant reveals the involvement of GABA transaminase in salt stress tolerance. BMC Plant Biol 2010; 10:20; http://dx.doi.org/10.1186/1471-2229-10-20; PMID: 20122158
- Renault H, El Amrani A, Berger A, Mouille G, Soubigou-Taconnat L, Bouchereau A, et al. γ-Aminobutyric acid transaminase deficiency impairs central carbon metabolism and leads to cell wall defects during salt stress in Arabidopsis roots. Plant Cell Environ 2013; PMID: 23148892
- Clark SM, Di Leo R, Dhanoa PK, Van Cauwenberghe OR, Mullen RT, Shelp BJ. Biochemical characterization, mitochondrial localization, expression, and potential functions for an Arabidopsis γ-aminobutyrate transaminase that utilizes both pyruvate and glyoxylate. J Exp Bot 2009; 60:1743 - 57; http://dx.doi.org/10.1093/jxb/erp044; PMID: 19264755
- Clark SM, Di Leo R, Van Cauwenberghe OR, Mullen RT, Shelp BJ. Subcellular localization and expression of multiple tomato γ-aminobutyrate transaminases that utilize both pyruvate and glyoxylate. J Exp Bot 2009; 60:3255 - 67; http://dx.doi.org/10.1093/jxb/erp161; PMID: 19470656
- Shimajiri Y, Ozaki K, Kainou K, Akama K. Differential subcellular localization, enzymatic properties and expression patterns of γ-aminobutyric acid transaminases (GABA-Ts) in rice (Oryza sativa). J Plant Physiol 2013; 170:196 - 201; http://dx.doi.org/10.1016/j.jplph.2012.09.007; PMID: 23122787
- Toufighi K, Brady SM, Austin R, Ly E, Provart NJ. The botany array resource: e-northerns, expression angling, and promoter analyses. Plant J 2005; 43:153 - 63; http://dx.doi.org/10.1111/j.1365-313X.2005.02437.x; PMID: 15960624
- Provart N, Zhu T.. A browser-based functional classification SuperViewer for Arabidopsis genomics. Curr Comp Mol Biol 2003; 2003:271 - 2
- Kornberg HL, Beevers H. The glyoxylate cycle as a stage in the conversion of fat to carbohydrate in castor beans. Biochim Biophys Acta 1957; 26:531 - 7; http://dx.doi.org/10.1016/0006-3002(57)90101-4; PMID: 13499412
- Kornberg HL, Madsen NB. Synthesis of C4-dicarboxylic acids from acetate by a glyoxylate bypass of the tricarboxylic acid cycle. Biochim Biophys Acta 1957; 24:651 - 3; http://dx.doi.org/10.1016/0006-3002(57)90268-8; PMID: 13436500
- Graham IA, Denby KJ, Leaver CJ. Carbon catabolite repression regulates glyoxylate cycle gene expression in cucumber. Plant Cell 1994; 6:761 - 72; PMID: 12244257
- Eastmond PJ, Germain V, Lange PR, Bryce JH, Smith SM, Graham IA. Postgerminative growth and lipid catabolism in oilseeds lacking the glyoxylate cycle. Proc Natl Acad Sci USA 2000; 97:5669 - 74; http://dx.doi.org/10.1073/pnas.97.10.5669; PMID: 10805817
- Cornah JE, Germain V, Ward JL, Beale MH, Smith SM. Lipid utilization, gluconeogenesis, and seedling growth in Arabidopsis mutants lacking the glyoxylate cycle enzyme malate synthase. J Biol Chem 2004; 279:42916 - 23; http://dx.doi.org/10.1074/jbc.M407380200; PMID: 15272001