Abstract
The developmental process of ripening is unique to fleshy fruits and a key factor in fruit quality. The tomato (Solanum lycopersicum) MADS-box transcription factor RIPENING INHIBITOR (RIN), one of the earliest-acting ripening regulators, is required for broad aspects of ripening, including ethylene-dependent and -independent pathways. However, our knowledge of direct RIN target genes has been limited, considering the broad effects of RIN on ripening. In a recent work published in The Plant Cell, we identified 241 direct RIN target genes by chromatin immunoprecipitation coupled with DNA microarray (ChIP-chip) and transcriptome analysis. Functional classification of the targets revealed that RIN participates in the regulation of many biological processes including well-known ripening processes such as climacteric ethylene production and lycopene accumulation. In addition, we found that ethylene is required for the full expression of RIN and several RIN-targeting transcription factor genes at the ripening stage. Here, based on our recently published findings and additional data, we discuss the ripening processes regulated by RIN and the interplay between RIN and ethylene.
In a developmental process unique to plant species bearing fleshy fruits, ripening drastically changes fruit characteristics including color, texture, aroma and nutritional composition. Ripening is thus a key step to determine fruit quality, and the understanding of the genetic program regulating fruit ripening will provide useful information to facilitate breeding of high quality fruit crops. In tomato (Solanum lycopersicum), ripening is regulated mainly by ethylene and several transcription factors (TFs).Citation1,Citation2 One of the earliest-acting TFs, the tomato MADS-box RIPENING INHIBITOR (RIN), is required for both ethylene-dependent and -independent ripening regulatory pathways.Citation2,Citation3 The rin mutation causes a severe inhibition of a broad range of ripening processes including softening, carotenoid accumulation and ripening-associated ethylene productionCitation4 (). This suggests that RIN acts as a master regulator of ripening and recent studies revealed that RIN may directly regulate the expression of two dozen ripening-associated genes.Citation5-Citation9 However, our knowledge of RIN target genes has been limited, considering the central role of RIN in regulation of ripening. In our recent work, published in The Plant Cell, we report a large-scale identification of direct RIN targets by chromatin immunoprecipitation coupled with DNA microarray (ChIP-chip) and transcriptome analysis.Citation10 In the report, we analyzed RIN binding to DNA from ripening tomato fruits by ChIP-chip using anti-RIN antibodies and a microarray for tomato gene promoters. This ChIP-chip analysis detected 1,046 RIN binding sites and, in combination with transcriptome data for wild type and rin mutant tomato fruits,Citation6 we identified 241 genes as direct RIN targets. These targets are positively or negatively regulated by RIN, generally contain a RIN binding site in their promoters, and include 11 known ripening-related genes.
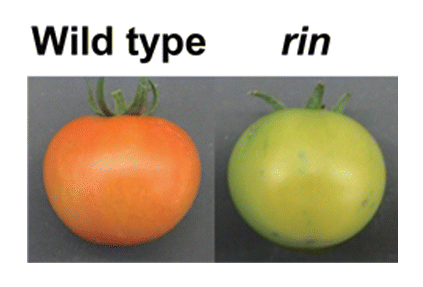
Figure 1. The rin mutation inhibits tomato fruit ripening. Wild type tomato fruits turn red and soften at the ripening stage (left), whereas the rin mutant fruits do not exhibit these changes (right). The rin mutation also inhibits the ripening-associated ethylene production and rise in respiration rate.Citation14 The induction of RIN at the onset of ripening is not influenced by exogenous ethylene.Citation3 In addition, the rin fruits show induction of some ethylene-responsive genes but do not ripen in response to exposure to exogenous ethylene,Citation14 suggesting that RIN is required for both ethylene-dependent and independent pathways.Citation2
Functional classification of the direct RIN targets with gene ontology (GO)
Our results in the recent paperCitation10 suggested that RIN participates in many biological pathways or processes during ripening, especially in ethylene production, lycopene accumulation and stress responses. For a more comprehensive interpretation of RIN involvement in ripening, we here provide an additional functional classification of the direct RIN targets based on similarity to Arabidopsis proteins with GO information.
In the positively regulated subset of direct RIN targets, GO analysis showed a significantly higher frequency of genes classified into categories related to ethylene synthesis (GO:0008898 and GO:0009693) than the frequency in the whole genome (). A similar enrichment was found in categories related to responses to biotic or abiotic stresses (GO:0009816, GO:0051707, GO:0009611, GO:0016165 and GO:0040007) (), which may reflect increased susceptibility of ripening fruit to pathogens.Citation1 These results are consistent with those with the Arabidopsis protein annotation database provided by the Munich Information Center for Protein Sequences (MIPS) as reported.Citation10 By contrast, it may seem strange that the GO analysis also found a significantly higher frequency in the category “Flower development” (GO:0009908) (). However, this reflects differences in function between tomato and Arabidopsis homologous genes, resulting in tomato genes involved in fruit ripening regulation, such as APETALA2a (AP2a), COLORLESS NON RIPENING (CNR) and RIN, being categorized in “Flower development” based on the annotation of their Arabidopsis homologs AP2, SQUAMOSA promoter binding protein and SEPALLATA genes. The implementation of functional gene annotation suitable for fleshy-fruit bearing plant species will enable more informative prediction of functional categories associated with RIN. This may help clarify the role of RIN-targeting genes or biological processesCitation10 for which the association with ripening remains unclear.
Table 1. Overrepresented GO categories in the direct RIN target genes
In the negatively regulated subset of direct RIN targets, GO analysis showed a significantly higher frequency in categories associated with TFs (GO:0003700 and GO:0010093) than the frequency in the whole genome (). This implies that RIN plays a role in irreversibly promoting ripening via the negative regulation of TF genes.
The interplay between RIN and ethylene might play a key role in fruit ripening regulation
In our recent paper,Citation10 we analyzed the ethylene responsiveness of TF genes directly and positively regulated by RIN, including NON RIPENING (NOR), CNR, TDR4, AP2a and RIN itself. The treatment with the ethylene signaling inhibitor 1-methylcyclopropene (1-MCP) substantially decreased the expression levels of RIN and eight TF genes, including NOR, TDR4 and AP2a, during ripening to 50% or less of the levels in the mock control. This indicated that the upregulation of RIN during ripening is affected by ethylene levels.
Based on our results, in the recent paper we proposed a model involving cooperative upregulation of the levels of RIN and ethylene during ripening.Citation10 In this model: (1) at the onset of fruit ripening, RIN is initially induced by an unknown developmental factor; (2) RIN induces ethylene production by upregulating genes for ethylene synthetic enzymes such as 1-aminocyclopropane-1-carboxylic acid (ACC) synthase (ACS2 and ACS4) and ACC oxidase (E8 and ACO6 directly, and ACO1 indirectly);Citation5,Citation8,Citation11 (3) RIN-induced and subsequent autocatalytic ethylene stimulates ethylene signaling pathways; (4) these signaling pathways, in turn, enhance RIN expression in a positive feedback loop. The expression of RIN is also enhanced by its direct autoregulation.Citation5 Thus, our model suggests that the full expression of RIN requires the ethylene-mediated feedback pathway in addition to the developmental controls. The increased RIN and ethylene might induce up- or downregulation of numerous ripening-related genes promptly at the early ripening stage, leading to synchronized initiation of ripening processes. Moreover, in our model, RIN modulates ethylene levels via the upregulation of AP2a, a negative regulator of ethylene production during ripening.Citation12,Citation13 This RIN-AP2a pathway might contribute to maintaining the ripening stage by regulating ethylene levels. In RIN and other RIN-targeted TF genes such as NOR and TDR4, ethylene was required for full induction of expression; thus it is possible that unknown ethylene-inducible factors may be involved in the induction of these genes. Identification of these unknown ethylene-inducible factors will provide a clue as to how the ripening-specific upregulation of RIN and other TFs is controlled.
| Abbreviations: | ||
| ACC | = | 1-aminocyclopropane-1-carboxylic acid |
| CArG box | = | C-A/T-rich-G box |
| ChIP-chip | = | chromatin immunoprecipitation coupled with DNA microarray |
| GO | = | gene ontology |
| 1-MCP | = | 1-methylcyclopropene |
| MIPS | = | the Munich Information Center for Protein Sequences |
| TF | = | transcription factor |
| RIN | = | RIPENING INHIBITOR |
| ChIP | = | chromatin immunoprecipitation |
Acknowledgment
This work was supported in part by the Program for Promotion of Basic and Applied Researches for Innovations in the Bio-oriented Technology Research Advancement Institution (BRAIN) of Japan to Y.I.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Giovannoni JJ. Genetic regulation of fruit development and ripening. Plant Cell 2004; 16:Suppl S170 - 80; http://dx.doi.org/10.1105/tpc.019158; PMID: 15010516
- Giovannoni JJ. Fruit ripening mutants yield insights into ripening control. Curr Opin Plant Biol 2007; 10:283 - 9; http://dx.doi.org/10.1016/j.pbi.2007.04.008; PMID: 17442612
- Vrebalov J, Ruezinsky D, Padmanabhan V, White R, Medrano D, Drake R, et al. A MADS-box gene necessary for fruit ripening at the tomato ripening-inhibitor (rin) locus. Science 2002; 296:343 - 6; http://dx.doi.org/10.1126/science.1068181; PMID: 11951045
- Tigchelaar EC, Mcglasson WB, Buescher RW. Genetic-regulation of tomato fruit ripening. HortScience 1978; 13:508 - 13
- Fujisawa M, Nakano T, Ito Y. Identification of potential target genes for the tomato fruit-ripening regulator RIN by chromatin immunoprecipitation. BMC Plant Biol 2011; 11:26; http://dx.doi.org/10.1186/1471-2229-11-26; PMID: 21276270
- Fujisawa M, Shima Y, Higuchi N, Nakano T, Koyama Y, Kasumi T, et al. Direct targets of the tomato-ripening regulator RIN identified by transcriptome and chromatin immunoprecipitation analyses. Planta 2012; 235:1107 - 22; http://dx.doi.org/10.1007/s00425-011-1561-2; PMID: 22160566
- Ito Y, Kitagawa M, Ihashi N, Yabe K, Kimbara J, Yasuda J, et al. DNA-binding specificity, transcriptional activation potential, and the rin mutation effect for the tomato fruit-ripening regulator RIN. Plant J 2008; 55:212 - 23; http://dx.doi.org/10.1111/j.1365-313X.2008.03491.x; PMID: 18363783
- Martel C, Vrebalov J, Tafelmeyer P, Giovannoni JJ. The tomato MADS-box transcription factor RIPENING INHIBITOR interacts with promoters involved in numerous ripening processes in a COLORLESS NONRIPENING-dependent manner. Plant Physiol 2011; 157:1568 - 79; http://dx.doi.org/10.1104/pp.111.181107; PMID: 21941001
- Qin G, Wang Y, Cao B, Wang W, Tian S. Unraveling the regulatory network of the MADS box transcription factor RIN in fruit ripening. Plant J 2012; 70:243 - 55; http://dx.doi.org/10.1111/j.1365-313X.2011.04861.x; PMID: 22098335
- Fujisawa M, Nakano T, Shima Y, Ito Y. A large-scale identification of direct targets of the tomato MADS box transcription factor RIPENING INHIBITOR reveals the regulation of fruit ripening. Plant Cell 2013; 25:371 - 86; http://dx.doi.org/10.1105/tpc.112.108118; PMID: 23386264
- Lin Z, Hong Y, Yin M, Li C, Zhang K, Grierson D. A tomato HD-Zip homeobox protein, LeHB-1, plays an important role in floral organogenesis and ripening. Plant J 2008; 55:301 - 10; http://dx.doi.org/10.1111/j.1365-313X.2008.03505.x; PMID: 18397374
- Chung MY, Vrebalov J, Alba R, Lee J, McQuinn R, Chung JD, et al. A tomato (Solanum lycopersicum) APETALA2/ERF gene, SlAP2a, is a negative regulator of fruit ripening. Plant J 2010; 64:936 - 47; http://dx.doi.org/10.1111/j.1365-313X.2010.04384.x; PMID: 21143675
- Karlova R, Rosin FM, Busscher-Lange J, Parapunova V, Do PT, Fernie AR, et al. Transcriptome and metabolite profiling show that APETALA2a is a major regulator of tomato fruit ripening. Plant Cell 2011; 23:923 - 41; http://dx.doi.org/10.1105/tpc.110.081273; PMID: 21398570
- Lincoln JE, Fischer RL. Regulation of gene expression by ethylene in wild-type and rin tomato (Lycopersicon esculentum) fruit. Plant Physiol 1988; 88:370 - 4; http://dx.doi.org/10.1104/pp.88.2.370; PMID: 16666310