Abstract
Plant development is dependent on the coordination between growth and cell proliferation. The nutrient sensing TOR kinase and its downstream target, the 40S ribosomal S6 Kinase, are central controllers of cell growth that were also shown to determine cell size by inhibiting the onset of mitosis in yeast and animal cells. We have shown that the Arabidopsis S6 Kinase1 inhibits cell proliferation through the RBR-E2FB complex. S6K1 interacts with RBR via its N-terminal RBR binding motif, promotes its nuclear localization and consequent RBR-dependent repression of cell cycle genes through E2FB. Here we show that S6K1 and E2FB are in a mutually antagonistic relationship both in their protein abundance and in their activity. We propose that this double inhibitory regulatory connection between S6K1 and E2FB forms a regulatory switch that might be important to determine whether cells divide or grow.
Growth and cell proliferation are intimately interlinked processes. In yeast, growth precedes and is required for cell proliferation, while in animal cells it is thought to be either upstream, or in parallel pathways that respond to specific stimuli such as growth or mitogenic factors.Citation1,Citation2 Nutrients and growth factors regulate the entry into cell proliferation by stimulating the accumulation of G1-phase specific cyclins in yeast, plant and animal cells. In addition, growth conditions can also affect the size of cells; yeast cells grown at favorable conditions are larger than when grown in poor media.Citation2-Citation4 The TOR signaling pathway is an evolutionary conserved nutrient sensor that sets the growth capacity of cells by regulating protein synthesis.Citation5-Citation7 It was shown that nutrient limiting conditions and inhibition of TOR activity, specifically the TORC1 complex, lead to the onset of mitosis and thus to reduced cell size both in yeast and in animal cells.Citation4,Citation8 Starvation activates the AMP-activated kinase (AMPK1) that was also shown to regulate mitosis directly or through the inhibition of TOR.Citation9-Citation11 Importantly, both upstream regulators and downstream effectors of the TORC1 complex, such as the 40S ribosomal protein S6 kinase were shown to be involved in the cell size control.Citation12-Citation14
In plant meristems, cell size homeostasis is also strictly controlled during cell proliferation. Similarly to animals, the TOR signaling pathway was shown to regulate growth, protein synthesis and metabolism in Arabidopsis.Citation15-Citation21 A conserved downstream TOR target is the Arabidopsis S6 Kinase (S6K) that phosphorylates the ribosomal protein S6.Citation21-Citation23 We have shown that S6K1 is a repressor of cell proliferation.Citation24 Interestingly, this function requires the RETINOBLASTOMA RELATED PROTEIN (RBR) and its target E2FB. S6K1 forms a complex with RBR and E2FB, promotes the nuclear localization of RBR and potentiates its repressor activity on E2FB targets, such as on the plant specific mitotic cyclin dependent kinase CDKB1;1 gene and on the S-phase specific ribonucleotide reductase RNR2 gene.Citation24 In this report we show that S6K1 and E2FB have an antagonistic relationship both at the protein abundance and activity levels. This double inhibition wiring in regulatory networks is an important feature of molecular switches that regulate cellular decisions.Citation25
Silencing of S6K1 Promotes E2FB Transcriptional Activity
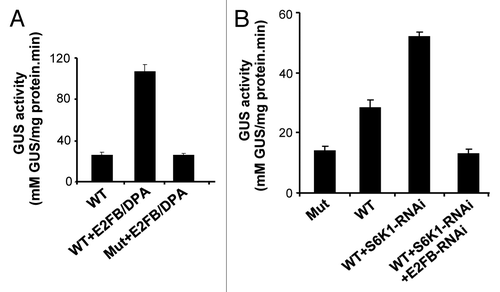
We have shown before that overexpression of the E2FB/DPA heterodimer induces the expression of CDKB1;1.Citation26 In agreement, transient overexpression of the E2FB/DPA heterodimer in protoplasts resulted in higher activity of the CDKB1;1 promoter, an effect abolished when the E2F-specific binding site was mutated (). We also found that depletion of S6K expression by RNA interference, resulted in an increased activity of the CDKB1;1 promoterCitation24 (). To demonstrate that S6K silencing affects CDKB1;1 promoter activity through E2FB, we co-silenced E2FB and S6K and found that the CDKB1;1 promoter activity was reduced.
Figure 1. S6K1 regulates the CDKB1;1 promoter activity through E2FB. (A) The CDKB1;1 WT promoter (WT) and CDKB1;1 mutant (Mut) promoter, where the consensus E2F binding site was mutatedCitation35 were fused to the GUS reporter gene and GUS activity measured in cells co-transformed with E2FB/DPA constructs. (B) Determination of activity of the CDKB1;1 WT and Mut promoter fused to the GUS reporter gene in cells transformed with a S6K1-RNAi construct alone or together with E2FB-RNAi.
Arabidopsis S6K1 and E2FB Oppositely Affect Each Other at the Post-Translational Level
We showed that S6K1, E2FB and RBR co-immunoprecipitated in vivo and thus are in a protein complex.Citation24 We investigated whether changes in the level of any of these proteins could affect the abundance of the complex components. Surprisingly, we found that silencing of S6K led to an increase of E2FB protein level, as detected through the Myc-epitope tagged construct of E2FB, expressed under the control of CaMV35S promoter in transformed protoplasts (). To demonstrate that S6K1 kinase activity is required for the regulation of E2FB protein level, we overexpressed a mutant, kinase-dead version of S6K1 that also resulted in increased E2FB levels (). In contrast to E2FB, S6K silencing had no effect on RBR protein level ().
Figure 2. Mutual inhibition of S6K1 and E2FB is regulated at the stability and activity levels. (A) S6K regulates E2FB accumulation. Detection of Myc-E2FB expressed from a construct transformed to control cells (first lane, upper panel) or to cells co-transformed with the S6K1-RNAi (second lane, upper panel). Myc-E2FB in control cells (first lane, lower panel), in cells co-transformed with S6K1-KD-HA, a kinase-dead form with the active center mutated (second lane, lower panel) and in cells co-transformed with S6K1-HA (third lane, lower panel). (B) RBR expressed from a construct transformed to control cells (first lane) or in cells co-transformed with the S6K1-RNAi construct (second lane). (C) RBR-E2FB regulate S6K1 accumulation. S6K1-HA expressed from a construct transformed to control cells (first lane), to cells co-transformed with the RBR-RNAi (second lane) or to cells co-transformed with E2FB-RNAi (third lane). (D) Regulation of E2FB and S6K stability. Upper panel: detection of Myc-E2FB expressed from a construct transformed to control cells or to cells co-transformed with the S6K1-RNAi construct in the presence of 100 μM of cycloheximide (CHX) at 0 and 6h. Lower panel: detection of S6K1-HA expressed from a construct transformed to control cells or to cells co-transformed with the E2FB-RNAi construct in the presence of 100 μM of CHX from 0 to 6h. (E) Phosphorylation of RBR-pocket-GST fusion protein (first row) and only GST (third row) with immunopurified S6K1-HA and S6K1-KD-HA expressed in control cells or in cells where E2FB was silenced by RNAi (E2FB-RNAi). Fifth row: detection of S6K1-HA in these samples. Note the elevated expression level of S6K1-HA when E2FB was silenced, a result that corresponds to (A). (F) Quantification of the phosphorylation signals shown in (D).
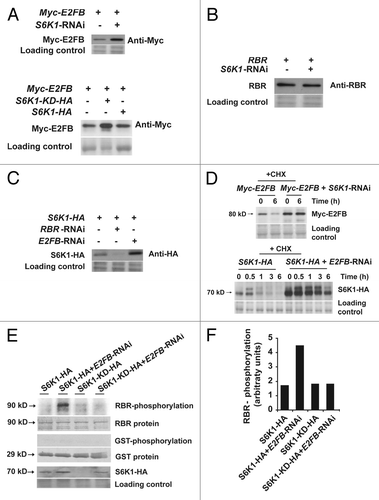
Next we investigated how E2FB and RBR levels affected S6K abundance by co-transformation of S6K1-HA with either E2FB-RNAi or RBR-RNAi constructs. When E2FB was silenced the S6K1-HA level increased. In contrast, RBR silencing led to a decrease in S6K1-HA ().
We also determined whether the E2FB and S6K1 levels are regulated through a change in protein stability. To do this, we compared Myc-E2FB protein levels in control and S6K silenced cells in conditions when the de novo protein synthesis was inhibited by cycloheximide (CHX). The E2FB level was elevated in S6K-silenced cells already at time 0, and this did not diminish in the presence of CHX, while in control cells the level of E2FB decreased (, upper panel). In a corresponding experiment, we found that silencing of E2FB led to an increased S6K1-HA level, which diminished slowly compared with the control cells (from less than 1h to around 6h) (, lower panel). This data suggested that the opposite function of E2FB and S6K on cell proliferation could be reinforced by their reciprocal effect on each others’ stability.
S6K1 Phosphorylates RBR Specifically When E2FB is Silenced
We have shown that S6K1 regulates RBR activity to repress the mitotic CDKB1;1 gene. We therefore investigated whether S6K could directly phosphorylate RBR and if this activity was regulated by the E2FB levels. To do this, we expressed in Arabidopsis cells a wild type S6K1-HA construct and a kinase-dead (S6K1-KD-HA) mutant version of it. We purified S6K1 through the HA-tag from Arabidopsis cells with and without E2FB silencing and assayed the in vitro phosphorylation of a GST-RBR-pocket substrate protein, a region that is responsible for binding to the E2F transcription factors. S6K1-HA, immunoprecipitated from control cells had undetectable phosphorylation activity on the RBR pocket region. In contrast, immunoprecipitation of S6K1-HA from cells co-transformed with an E2FB-RNAi construct resulted in an active S6K1-HA that phosphorylated the GST-RBR-pocket substrate (). To demonstrate that S6K1 kinase activity, and not another co-purified kinase, is responsible for RBR phopshorylation, we also transformed a kinase-dead mutant form of S6K1 (S6K1-KD-HA) which did not phosphorylate the GST-RBR pocket fragment, even when E2FB was silenced (). Interestingly, the kinase-dead form of S6K1 was much less abundant than the active S6K1 (). These results indicate that S6K1 regulates RBR by direct phosphorylation specifically when E2FB levels are low.
In our previous work we showed that S6K1 negatively regulates cell proliferation by promoting the cell cycle inhibitory function of the plant RB ortholog, RBR.Citation24 A similar effect was recently reported when S6K1 was expressed under the control of the auxin responsive element DR5.Citation27 We have shown that S6K1 interacts with RBR and E2FB leading to E2FB repression. In agreement, S6K silencing increases the activity of E2FB target genes, including the plant specific mitotic CDKB1;1.Citation26 Recently, we have shown that sucrose levels control the amount and activity of RBR/E2Fs complexes, leading to different outcomes in cell growth and proliferation.Citation28 In the presence of sucrose RBR is phosphorylated, dependent on CYCD3;1 and KRP2 levels (). This inactivates RBR, leading to the release of E2FB that will stimulate cell proliferation. In agreement, overexpression of E2FB accelerates cell cycle progression and cells divide with a smaller cell size.Citation26,Citation29 The dramatic effect of E2FB overexpression on total cell mass also indicated an inhibition of cell growth but the underlying molecular mechanisms were not known.Citation26 Here we show that S6K1 and E2FB are in a mutually antagonistic relationship both in their protein abundance and in their activity. Silencing of E2FB activates S6K1 that phosphorylates RBR, while silencing of S6K1 leads to more E2FB that can trans-activate the mitotic regulator, CDKB1;1. The mutual inhibition of E2FB and S6K1 is regulated by protein stability, as we showed that S6K1 protein was stabilized in E2FB silenced cells, and E2FB became stabilized when S6K1 was silenced (). These observations seem to depend on E2FB and S6K1 activities. Overexpression of a dominant negative kinase-inactive S6K1 mutant construct also resulted in an increased E2FB protein level; while silencing of RBR, which releases active E2FB, destabilized the S6K1 protein (). The mechanism how E2FB and S6K regulate each other`s protein stability is not yet known. There are several candidate F-box protein genes carrying E2F-binding elements in their promotersCitation30 that could be induced by elevated E2FB levels and regulate protein stability. RBR, E2FA and E2FB are known to be unstable especially under sucrose-depleted conditions.Citation31,Citation32 S6K might regulate E2FB turnover by phosphorylation either directly or indirectly. This post-translational modification could then be a mark for proteasomal degradation similarly to E2FC and DPB.Citation33,Citation34
Figure 3. Model explaining the antagonistic functions of E2FB and S6K1. Sucrose availability increases the CycD3;1 amount and presumably the TOR kinase activity, and decreases KRP2. CycD3;1 in complex with CDKA phosphorylates RBR, which releases E2FB and that drives cells into the mitotic cell cycle. On one hand E2FB destabilizes S6K1 and negatively affect cell growth. On the other hand active S6K1 destabilizes E2FB and potentiates RBR to inhibit cell proliferation. This antagonistic relationship of S6K1 and E2FB could play an important role to keep cell proliferation as is the case of small meristematic cells, or allow cells to grow and increase the cell size.
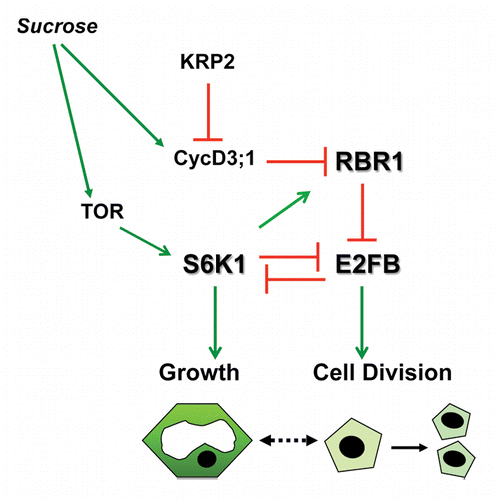
Collectively, these findings are consistent with a model that S6K1 represses cell proliferation by enhancing RBR function through several routes: by the regulation of RBR localization, by enhancing RBR binding to E2FB and by repressing E2FB accumulation. On the other hand accumulation of active E2FB represses cell growth through destabilization of S6K1. These opposing functions could provide the molecular link between cell growth (S6K) and cell proliferation (E2FB) ().
Materials and Methods
Constructs
The constructs used in this work have been described elsewhere, E2FB-RNAi,Citation28 S6K1-RNAi, S6K1-HA,Citation24 E2FB/DPA,Citation26 pRT100::S6K1-KD-HA (kinase-dead S6K1 where Lys163, responsible for phosphate coordination in the active center, was replaced by Arg) was kindly provided by Dr M. Teige, and confirmed by sequencing. E2FB was cloned in frame with the N-terminal double Myc epitope,Citation36 and the Myc tagged E2FB was further cloned into the pK7WG2D Gateway vector (Invitrogene) following the manufacturer’s instructions.
In vitro RBR kinase assay and tmmunoblotting
S6K1 activity toward the RBR-pocket region (RBR-PR) was measured in protoplasts transformed with S6K1-HA, S6K1-HA+E2FB-RNAi, S6K1-kinase dead (KD)-HA and S6K1-KD-HA+E2FB-RNAi constructs. HA antibody was used to immunoprecipitate S6K1 and kinase assays were performed as described.Citation37 The pGEX::RBR-PR polypeptide was purified accordingly to the manufacturers instructions (Promega) and 1 µg was used as substrate. Reactions were resolved in a 10% SDS-PAGE gel and the phosphorylation signal was detected on a Typhoon 9410 phosphorimager and quantified by the ImageQuant software (GE Healthcare). Immunoblotting was performed as described.Citation24
Protein stability assays
Protoplast cells were transformed with HA-S6K1 or Myc-E2FB constructs alone or in combination with E2FB-RNAi or S6K1-RNAi constructs, respectively and treated further with 100 μM of cycloheximide (CHX) for up to six hours and protein levels were followed by western blot analyses by using specific antibodies against the epitope tag as described in reference Citation24.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Conlon I, Raff M. Size control in animal development. Cell 1999; 96:235 - 44; http://dx.doi.org/10.1016/S0092-8674(00)80563-2; PMID: 9988218
- Jorgensen P, Tyers M. How cells coordinate growth and division. Curr Biol 2004; 14:R1014 - 27; http://dx.doi.org/10.1016/j.cub.2004.11.027; PMID: 15589139
- Davie E, Petersen J. Environmental control of cell size at division. Curr Opin Cell Biol 2012; 24:838 - 44; http://dx.doi.org/10.1016/j.ceb.2012.08.003; PMID: 22947494
- Petersen J, Nurse P. TOR signalling regulates mitotic commitment through the stress MAP kinase pathway and the Polo and Cdc2 kinases. Nat Cell Biol 2007; 9:1263 - 72; http://dx.doi.org/10.1038/ncb1646; PMID: 17952063
- Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol 2009; 10:307 - 18; http://dx.doi.org/10.1038/nrm2672; PMID: 19339977
- Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell 2006; 124:471 - 84; http://dx.doi.org/10.1016/j.cell.2006.01.016; PMID: 16469695
- Robaglia C, Thomas M, Meyer C. Sensing nutrient and energy status by SnRK1 and TOR kinases. Curr Opin Plant Biol 2012; 15:301 - 7; http://dx.doi.org/10.1016/j.pbi.2012.01.012; PMID: 22305521
- Fingar DC, Salama S, Tsou C, Harlow E, Blenis J. Mammalian cell size is controlled by mTOR and its downstream targets S6K1 and 4EBP1/eIF4E. Genes Dev 2002; 16:1472 - 87; http://dx.doi.org/10.1101/gad.995802; PMID: 12080086
- Banko MR, Allen JJ, Schaffer BE, Wilker EW, Tsou P, White JL, et al. Chemical genetic screen for AMPKα2 substrates uncovers a network of proteins involved in mitosis. Mol Cell 2011; 44:878 - 92; http://dx.doi.org/10.1016/j.molcel.2011.11.005; PMID: 22137581
- Hardie DG, Ross FA, Hawley SA. AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nat Rev Mol Cell Biol 2012; 13:251 - 62; http://dx.doi.org/10.1038/nrm3311; PMID: 22436748
- Robitaille AM, Hall MN. Ramping up mitosis: an AMPKα2-regulated signaling network promotes mitotic progression. Mol Cell 2012; 45:8 - 9; http://dx.doi.org/10.1016/j.molcel.2011.12.018; PMID: 22244327
- Kozma SC, Thomas G. Regulation of cell size in growth, development and human disease: PI3K, PKB and S6K. Bioessays 2002; 24:65 - 71; http://dx.doi.org/10.1002/bies.10031; PMID: 11782951
- Montagne J, Stewart MJ, Stocker H, Hafen E, Kozma SC, Thomas G. Drosophila S6 kinase: a regulator of cell size. Science 1999; 285:2126 - 9; http://dx.doi.org/10.1126/science.285.5436.2126; PMID: 10497130
- Shima H, Pende M, Chen Y, Fumagalli S, Thomas G, Kozma SC. Disruption of the p70(s6k)/p85(s6k) gene reveals a small mouse phenotype and a new functional S6 kinase. EMBO J 1998; 17:6649 - 59; http://dx.doi.org/10.1093/emboj/17.22.6649; PMID: 9822608
- Deprost D, Yao L, Sormani R, Moreau M, Leterreux G, Nicolaï M, et al. The Arabidopsis TOR kinase links plant growth, yield, stress resistance and mRNA translation. EMBO Rep 2007; 8:864 - 70; http://dx.doi.org/10.1038/sj.embor.7401043; PMID: 17721444
- Dobrenel T, Marchive C, Sormani R, Moreau M, Mozzo M, Montané MH, et al. Regulation of plant growth and metabolism by the TOR kinase. Biochem Soc Trans 2011; 39:477 - 81; http://dx.doi.org/10.1042/BST0390477; PMID: 21428923
- Menand B, Desnos T, Nussaume L, Berger F, Bouchez D, Meyer C, et al. Expression and disruption of the Arabidopsis TOR (target of rapamycin) gene. Proc Natl Acad Sci USA 2002; 99:6422 - 7; http://dx.doi.org/10.1073/pnas.092141899; PMID: 11983923
- Moreau M, Azzopardi M, Clément G, Dobrenel T, Marchive C, Renne C, et al. Mutations in the Arabidopsis homolog of LST8/GβL, a partner of the target of Rapamycin kinase, impair plant growth, flowering, and metabolic adaptation to long days. Plant Cell 2012; 24:463 - 81; http://dx.doi.org/10.1105/tpc.111.091306; PMID: 22307851
- Ren M, Venglat P, Qiu S, Feng L, Cao Y, Wang E, et al. Target of rapamycin signaling regulates metabolism, growth, and life span in Arabidopsis.. Plant Cell 2012; 24:4850 - 74; http://dx.doi.org/10.1105/tpc.112.107144; PMID: 23275579
- John F, Roffler S, Wicker T, Ringli C. Plant TOR signaling components. Plant Signal Behav 2011; 6:1700 - 5; http://dx.doi.org/10.4161/psb.6.11.17662; PMID: 22057328
- Xiong Y, Sheen J. Rapamycin and glucose-target of rapamycin (TOR) protein signaling in plants. J Biol Chem 2012; 287:2836 - 42; http://dx.doi.org/10.1074/jbc.M111.300749; PMID: 22134914
- Mahfouz MM, Kim S, Delauney AJ, Verma DPS. Arabidopsis TARGET OF RAPAMYCIN interacts with RAPTOR, which regulates the activity of S6 kinase in response to osmotic stress signals. Plant Cell 2006; 18:477 - 90; http://dx.doi.org/10.1105/tpc.105.035931; PMID: 16377759
- Turck F, Zilbermann F, Kozma SC, Thomas G, Nagy F. Phytohormones participate in an S6 kinase signal transduction pathway in Arabidopsis.. Plant Physiol 2004; 134:1527 - 35; http://dx.doi.org/10.1104/pp.103.035873; PMID: 15064379
- Henriques R, Magyar Z, Monardes A, Khan S, Zalejski C, Orellana J, et al. Arabidopsis S6 kinase mutants display chromosome instability and altered RBR1-E2F pathway activity. EMBO J 2010; 29:2979 - 93; http://dx.doi.org/10.1038/emboj.2010.164; PMID: 20683442
- Tyson JJ, Novak B. Cell cycle: who turns the crank?. Curr Biol 2011; 21:R185 - 7; http://dx.doi.org/10.1016/j.cub.2011.01.042; PMID: 21377093
- Magyar Z, De Veylder L, Atanassova A, Bakó L, Inzé D, Bögre L. The role of the Arabidopsis E2FB transcription factor in regulating auxin-dependent cell division. Plant Cell 2005; 17:2527 - 41; http://dx.doi.org/10.1105/tpc.105.033761; PMID: 16055635
- Shin YJ, Kim S, Du H, Choi S, Verma DP, Cheon C-I. Possible dual regulatory circuits involving AtS6K1 in the regulation of plant cell cycle and growth. Mol Cells 2012; 33:487 - 96; http://dx.doi.org/10.1007/s10059-012-2275-4; PMID: 22526395
- Magyar Z, Horváth B, Khan S, Mohammed B, Henriques R, De Veylder L, et al. Arabidopsis E2FA stimulates proliferation and endocycle separately through RBR-bound and RBR-free complexes. EMBO J 2012; 31:1480 - 93; http://dx.doi.org/10.1038/emboj.2012.13; PMID: 22307083
- De Veylder L, Beeckman T, Beemster GT, de Almeida Engler J, Ormenese S, Maes S, et al. Control of proliferation, endoreduplication and differentiation by the Arabidopsis E2Fa-DPa transcription factor. EMBO J 2002; 21:1360 - 8; http://dx.doi.org/10.1093/emboj/21.6.1360; PMID: 11889041
- Ramirez-Parra E, Fründt C, Gutierrez C. A genome-wide identification of E2F-regulated genes in Arabidopsis.. Plant J 2003; 33:801 - 11; http://dx.doi.org/10.1046/j.1365-313X.2003.01662.x; PMID: 12609051
- Hirano H, Shinmyo A, Sekine M. Both negative and positive G1 cell cycle regulators undergo proteasome-dependent degradation during sucrose starvation in Arabidopsis . Plant Signal Behav 2011; 6:1394 - 6; http://dx.doi.org/10.4161/psb.6.9.16877; PMID: 22019639
- Hirano H, Shinmyo A, Sekine M. Arabidopsis G1 cell cycle proteins undergo proteasome-dependent degradation during sucrose starvation. Plant Physiol Biochem 2011; 49:687 - 91; http://dx.doi.org/10.1016/j.plaphy.2011.03.001; PMID: 21444209
- del Pozo JC, Boniotti MB, Gutierrez C. Arabidopsis E2Fc functions in cell division and is degraded by the ubiquitin-SCF(AtSKP2) pathway in response to light. Plant Cell 2002; 14:3057 - 71; http://dx.doi.org/10.1105/tpc.006791; PMID: 12468727
- del Pozo JC, Diaz-Trivino S, Cisneros N, Gutierrez C. The balance between cell division and endoreplication depends on E2FC-DPB, transcription factors regulated by the ubiquitin-SCFSKP2A pathway in Arabidopsis.. Plant Cell 2006; 18:2224 - 35; http://dx.doi.org/10.1105/tpc.105.039651; PMID: 16920782
- Boudolf V, Vlieghe K, Beemster GTS, Magyar Z, Torres Acosta JA, Maes S, et al. The plant-specific cyclin-dependent kinase CDKB1;1 and transcription factor E2Fa-DPa control the balance of mitotically dividing and endoreduplicating cells in Arabidopsis.. Plant Cell 2004; 16:2683 - 92; http://dx.doi.org/10.1105/tpc.104.024398; PMID: 15377755
- Magyar Z, Atanassova A, De Veylder L, Rombauts S, Inzé D. Characterization of two distinct DP-related genes from Arabidopsis thaliana.. FEBS Lett 2000; 486:79 - 87; http://dx.doi.org/10.1016/S0014-5793(00)02238-9; PMID: 11108847
- Anthony RG, Henriques R, Helfer A, Mészáros T, Rios G, Testerink C, et al. A protein kinase target of a PDK1 signalling pathway is involved in root hair growth in Arabidopsis.. EMBO J 2004; 23:572 - 81; http://dx.doi.org/10.1038/sj.emboj.7600068; PMID: 14749726