Abstract
Post-translational modifications of core histones are important for various DNA-templated processes such as transcription and repair. We recently reported that the ALFIN LIKE 6 (AL6) gene, identified in a forward genetic screen, is critical for phosphate deficiency-induced root hair formation and several other processes associated with the regulation of cellular phosphate homeostasis. AL6 contains a Plant Homeo Domain (PHD) finger that can bind to trimethylated lysine 4 of histone H3 (H3K4me3). Homozygous mutants defective in AL6 expression form very short root hairs under phosphate-deficient conditions, presumably caused by altered expression of putative primary and secondary down-stream targets of AL6. In this Addendum, we speculate about possible roles of AL6, H3K4 trimethylation and other chromatin modifications in the adaptation of plants to low phosphate availability.
Phosphorous, mainly taken up as phosphate (Pi) by plants, is an essential mineral nutrient required as a constituent of macromolecular structures such as nucleic acids and phospholipids and for many vital processes including signaling and energy metabolism. Due to covalent and non-covalent interactions of Pi with metal ions and other soil constituents, Pi is extremely immobile and often not available in adequate amounts to support optimal plant growth. To improve the acquisition of Pi from immobile pools and to recalibrate cellular Pi homeostasis under conditions of low Pi availability, plants have evolved strategies that improve the acquisition, import and remobilization of Pi. As part of such strategies, Pi-deficient plants form longer and denser root hairs.Citation1,Citation2 The mechanisms underlying these phenotypic alterations are only partly understood. We recently described a mutant that is defective in Pi deficiency-induced root hair development, but forms normal hairs under Pi-replete conditions.Citation3 This mutant, named per2 (phosphate deficiency root hair defective 2), harbors a mutation in the ALFIN LIKE 6 (AL6) transcription factor, a member of the Alfin1-like homeodomain protein family. The PER2 mutation decreased the expression of several Pi-responsive genes, namely the Myb-type transcription factor ETC1, the non-specific phospholipase NPC4, the UDP-glycosyltransferase SQD2, and the Pi pyrophosphate-specific phosphatase PS2. Mutants harboring defects in these four genes showed root hair defects particularly under Pi-deficient conditions that partly resembled the per2 phenotype, suggesting that these genes are putative downstream targets of AL6. How AL6 regulates these genes remained unclear.
H3K4 Trimethylation Marks Actively Transcribed Genes
Transcriptional activation in response to environmental cues is an orchestrated action of the general transcriptional machinery, trans-acting factors, co-activators, enhancers and proteins that control postinitiation processes and pre-mRNA processing. Further, transcription and other DNA-templated processes are affected by the accessibility of DNA, which is modulated by post-translational modifications (PTMs), such as acetylation and methylation, of the amino terminal tails of histones. Methylation of histone tails is complex, since both the position of the modified residue and the extent of methylation (i.e., mono-, di-, or trimethylation) can alter gene activity. Generally, trimethylation of histone H3 on lysine 4 (H3K4me3) is associated with the 5′ region of transcriptionally active loci.Citation4,Citation5 However, H3K4me3 per se does not appear to influence transcription.Citation6,Citation7 The role of H3K4me3 in gene activation was rather described in terms of providing a binding platform for transcriptional activators and for factors that mediate transcript elongation and mRNA maturation.Citation4,Citation8
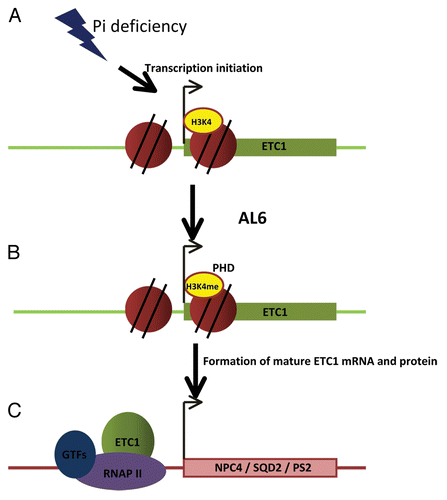
Recruitment of AL6 by H3K4me3 is Critical for the Phenotypic Readout
The language of post-translational histone modifications requires a “reader” to interpret the information which is then translated into gene activity. This translation is not categorical for a certain epigenetic mark and additional information encrypted in the context of other chromatin modifications and provided by proteins interacting with the readers or effector proteins is required for the correct interpretation of the code. AL6 is a bona fide reader that can bind to di- and, more readily, trimethylated H3K4.Citation9 Alfin proteins are plant-specific, but highly similar to the broadly distributed ING (inhibitor of growth) proteins and bromodomain PHD finger transcription factors.Citation10-Citation13 AL6 is the first AL or plant ING protein for which a function has been assigned, but the exact mechanism of how AL6 triggers the activation of multiple genes with important functions in cellular Pi homeostasis remains elusive. A working model describing a possible scenario is shown in . Among the putative AL6 targets inferred from transcriptional profiling experiments, etc1 mutants showed the most pronounced phenotype. In addition, etc1 plants showed de-regulated expression of NPC4, SQD2 and PS2, suggesting that ETC1 is controlling the expression of these genes. In roots of Pi-deficient plants, ETC1 is robustly expressed, a prerequisite for H3K4 trimethylation at the promoter-proximal nucleosomes of the gene. AL6 binds to lysine 4-trimethylated histone H3 through its PHD domain and thereby positively affects post-initiation events such as pre-mRNA maturation. Consequently, ETC1 binds to the promoters of NPC4, SQD2 and PS2 and activate the expression of these and other genes that support root hair elongation and other adaptive processes during Pi deficiency. How this process is regulated by Pi deficiency, however, remains unclear. The expression of AL6 is not affected by the Pi supply,Citation3 ruling out a simple transcriptional control. Global changes in histone methylation patterns in response to Pi deficiency have not been yet investigated, but dramatic changes in histone PTMs are unlikely to occur. In a survey on genome-wide changes in histone H3 lysine 4 methylation patterns in Arabidopsis seedlings subjected to dehydration, the H3K4me3 mark was mainly associated with genes but the localization was robust and did not change during the stress.Citation5 If the histone marks are not changing during Pi deficiency stress and the level of AL6 is also unchanged, how is the recruitment of AL6 regulated? An attractive scenario is that other histone PTMs are important to interpret the context of the code. Histone H3 trimethlated on lysine 4 undergoes dynamic acetylation in mammals and Dictyostelium.Citation14,Citation15 In Arabidopsis, salt stress induced H3K4 trimethylation and decreased the repressive mark H3K9me2 at abiotic stress-responsive genes.Citation16 H3K4 trimethylation was absent in the histone deacetylase 6 (HDA6) mutant axe1-5, suggesting that HAD activity is required for H3K4 methylation.Citation16 Together these data suggest that histone marks cannot be seen isolated but may act dynamically and synergistically to control transcription and post-transcriptional processes which, in turn, dictate gene activity. While such a scenario awaits further experimental exploration, it appears that together with transcriptomic and proteomic changes, additional layers of gene regulation, hidden in the dynamic language of histone PTMs, provide epigenetic strategies for the adaptation of plants to a constantly changing environment.
Figure 1. Hypothetical model of the molecular mechanism by which AL6 controls root hair elongation under low phosphate conditions. Phosphate deficiency triggers the initiation of active transcription of ETC1 (A). This leads to histone H3 lysine 4 trimethylation of the promoter-proximal elements of ETC1 followed by recognition and binding of AL6 via its PHD domain to H3K4me3 and formation of mature ETC1 mRNA (B). ETC1 might facilitate transcription of NPC4, SQD2 and PS2 (C).
Acknowledgments
This work was supported by grants from Academia Sinica. We thank the Arabidopsis Biological Resource Center (Ohio State University) for providing the T-DNA insertion mutants and wild-type seeds used in this study. We also thank Marjori Matzke (IPMB, Taiwan) for valuable suggestions and critical comments on the manuscript, Yuki Nakamura (IPMB, Taiwan) for providing npc4-1 and npc4-2 mutant seeds and Christoph Benning (Michigan State University) for providing sqd2 mutant seeds.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Ma Z, Bielenberg DG, Brown KM, Lynch JP. Regulation of root hair density by phosphorus availability in Arabidopsis thaliana. Plant Cell Environ 2001; 24:459 - 67; http://dx.doi.org/10.1046/j.1365-3040.2001.00695.x
- Müller M, Schmidt W. Environmentally induced plasticity of root hair development in Arabidopsis. Plant Physiol 2004; 134:409 - 19; http://dx.doi.org/10.1104/pp.103.029066; PMID: 14730071
- Chandrika NN, Sundaravelpandian K, Yu SM, Schmidt W. ALFIN-LIKE 6 is involved in root hair elongation during phosphate deficiency in Arabidopsis. New Phytol 2013; In press http://dx.doi.org/10.1111/nph.12194; PMID: 23432399
- Sims RJ 3rd, Millhouse S, Chen CF, Lewis BA, Erdjument-Bromage H, Tempst P, et al. Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Mol Cell 2007; 28:665 - 76; http://dx.doi.org/10.1016/j.molcel.2007.11.010; PMID: 18042460
- van Dijk K, Ding Y, Malkaram S, Riethoven JJ, Liu R, Yang J, et al. Dynamic changes in genome-wide histone H3 lysine 4 methylation patterns in response to dehydration stress in Arabidopsis thaliana.. BMC Plant Biol 2010; 10:238; http://dx.doi.org/10.1186/1471-2229-10-238; PMID: 21050490
- Pavri R, Zhu B, Li GH, Trojer P, Mandal S, Shilatifard A, et al. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell 2006; 125:703 - 17; http://dx.doi.org/10.1016/j.cell.2006.04.029; PMID: 16713563
- Sims RJ 3rd, Reinberg D. Histone H3 Lys 4 methylation: caught in a bind?. Genes Dev 2006; 20:2779 - 86; http://dx.doi.org/10.1101/gad.1468206; PMID: 17043307
- Wysocka J, Swigut T, Milne TA, Dou YL, Zhang X, Burlingame AL, et al. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell 2005; 121:859 - 72; http://dx.doi.org/10.1016/j.cell.2005.03.036; PMID: 15960974
- Lee WY, Lee D, Chung WI, Kwon CS. Arabidopsis ING and Alfin1-like protein families localize to the nucleus and bind to H3K4me3/2 via plant homeodomain fingers. Plant J 2009; 58:511 - 24; http://dx.doi.org/10.1111/j.1365-313X.2009.03795.x; PMID: 19154204
- Peña PV, Davrazou F, Shi X, Walter KL, Verkhusha VV, Gozani O, et al. Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2. Nature 2006; 442:100 - 3; PMID: 16728977
- Shi X, Kachirskaia I, Walter KL, Kuo JH, Lake A, Davrazou F, et al. Proteome-wide analysis in Saccharomyces cerevisiae identifies several PHD fingers as novel direct and selective binding modules of histone H3 methylated at either lysine 4 or lysine 36. J Biol Chem 2007; 282:2450 - 5; http://dx.doi.org/10.1074/jbc.C600286200; PMID: 17142463
- Aasland R, Gibson TJ, Stewart AF. The PHD finger: implications for chromatin-mediated transcriptional regulation. Trends Biochem Sci 1995; 20:56 - 9; http://dx.doi.org/10.1016/S0968-0004(00)88957-4; PMID: 7701562
- Bienz M. The PHD finger, a nuclear protein-interaction domain. Trends Biochem Sci 2006; 31:35 - 40; http://dx.doi.org/10.1016/j.tibs.2005.11.001; PMID: 16297627
- Hazzalin CA, Mahadevan LC. Dynamic acetylation of all lysine 4-methylated histone H3 in the mouse nucleus: analysis at c-fos and c-jun. PLoS Biol 2005; 3:e393; http://dx.doi.org/10.1371/journal.pbio.0030393; PMID: 16262446
- Hsu DW, Chubb JR, Muramoto T, Pears CJ, Mahadevan LC. Dynamic acetylation of lysine-4-trimethylated histone H3 and H3 variant biology in a simple multicellular eukaryote. Nucleic Acids Res 2012; 40:7247 - 56; http://dx.doi.org/10.1093/nar/gks367; PMID: 22600736
- Chen LT, Luo M, Wang YY, Wu KQ. Involvement of Arabidopsis histone deacetylase HDA6 in ABA and salt stress response. J Exp Bot 2010; 61:3345 - 53; http://dx.doi.org/10.1093/jxb/erq154; PMID: 20519338