Abstract
The GARP domain is a single Myb-related DNA-binding domain found in plant transcription factors. Proteins containing the GARP domain (GARP family proteins) are suggested to be involved in the regulation of various physiological processes through their interactions with ostensibly different DNA sequences. Our recent study on a nitrate-inducible gene encoding a GARP family protein, referred to as NIGT1 (Nitrate-Inducible, GARP-type Transcriptional Repressor 1), not only suggests a previously unidentified role for the GARP family proteins in higher plants but also provides a hypothesis for why NIGT1 can show dual specificity on DNA binding and why respective GARP family proteins can recognize very different DNA sequences.
The GARP domain is a single Myb-related DNA-binding domain, which was named using the initials of the GARP domain-containing proteins, GOLDEN2, ARR-B and Psr1.Citation1 Many genes in the plant genomes encode GARP family proteins, which are likely to be involved in the regulation of a variety of physiological processes. In the case of Arabidopsis, more than 50 genes have been estimated to encode GARP family proteins.Citation2
In previous studies, physiological roles of several GARP family proteins have been investigated. GOLDEN2 is a maize protein that functions as a transcriptional regulator of cellular differentiation in leaves,Citation3 whereas ARR-B proteins are type-B response regulators involved in His-Asp phospho-relay signal transduction systems.Citation4 Psr1 is a protein involved in phosphate starvation signaling in Chlamydomonas.Citation5 Other characterized GARP family proteins in Arabidopsis are PHR1 that is associated with a phosphate starvation response,Citation6 GLK1 and GLK2 that are two closely related members involved in the synchronized expression of nuclear photosynthetic genesCitation7,Citation8 and PCL1 which is essential for generating clock oscillation.Citation9
The results of DNA-binding experiments in previous studies have revealed that respective GARP family proteins bind to different DNA sequences in a sequence-specific manner. Structural analysis of an Arabidopsis ARR-B protein, ARR10, by NMR spectroscopy has shown that its GARP domain contains a helix-turn-helix structure and binds to an AGATT sequence.Citation10 Through comparisons of the DNA sequences of the target promoters of GLK1 and GLK2, the binding sequence for these two GARP proteins was suggested to be CCAATC.Citation8 The ARR10- and GLK1/2-binding sequences are similar as they both contain an AATC sequence (in the case of ARR10, this is present in the complementary strand). On the other hand, PHR1 binds to GNATATNC, which is very different from the binding sequences for ARR10 and GLK1/2. Hence, the PHR1-binding sequence and ARR10- and GLK1/2-binding sequences have no common features, and respective GARP domains seem therefore to recognize very divergent sequences.
Our recent characterization of a rice nitrate-inducible gene encoding a GARP family protein, NIGT1 (Nitrate-Inducible, GARP-type Transcriptional Repressor 1), revealed that the product of this gene is a transcriptional repressor that regulates its own expression through direct binding to several sites within its own promoter. Furthermore, analyses of transgenic rice plants overexpressing this gene suggested that NIGT1 might play a role in modulating chlorophyll contents. Because the Arabidopsis homologs of rice NIGT1 are also nitrate-inducible genes and NIGT1-binding sites were found to be conserved in the promoter regions of the Arabidopsis homologs, we have proposed that NIGT1 and its homologs may play a similar role in nitrogen response in both monocots and dicots.Citation11
In addition to the implications of the identified physiological role of the GARP family proteins in the plant kingdom, our characterization of NIGT1 as a DNA-binding protein also provided a new insight that may explain why respective GARP family proteins recognize such different DNA sequences. The results of our binding experiments using NIGT1 indicated that it binds to two different sequences, GAATC and GAATATTC, which resemble the ARR10-binding sequence (AATCT) and the PHR1-binding sequence (GNATATNC), respectively. ARR10 binds to its target DNA as a monomer,Citation10 while PHR1 binds to its target palindromic sequence as a dimerCitation6 (). Considering that GAATATTC is a palindromic sequence with similarity to GAATC, I propose a hypothesis that may explain the dual specificity of NIGT1 in terms of DNA-binding ().
Figure 1. Models for the DNA binding of GARP family proteins. (A) Binding of ARR10, GLK, GLK2 and PHR1. (B) Binding of NIGT1 to GAATC and GAATATTC sequences. Amino acid residues of ARR10 that interact with DNA are indicated in black. Where corresponding amino acids are conserved between GLK1, GLK2, PHR1 and NIGT1, these residues are indicated in black. When corresponding amino acids are not conserved in these four GARP proteins, the residues are indicated in red. Amino residues that interact with the same base pair are indicated side by side. Positions of residues in GLK1/2 are shown according to the amino acid sequence of GLK1. Binding sequences of respective GARP family proteins are indicated by filled blue circles, and the nucleotides flanking these binding sequences are indicated by unfilled circles.
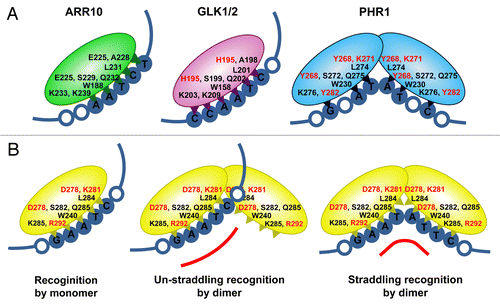
In this hypothesis, NIGT1 binds to GAATC as a monomer or through one subunit of a dimer, whereas it binds to GAATATTC though the interaction of both subunits of a dimer. In the binding to GAATC, the K285, W240, S282, Q285 and L284 residues of NIGT1 may participate in the interaction with the AATC base pairs, because the corresponding residues of ARR10 have been shown to do so. In the case of ARR10, the E225 and A228 residues interact with the last “T” in the AATCT site (). Consistent with the fact that the NIGT1-binding sequence does not include this “T,” the corresponding residues are D278 and K281 in NIGT1. On the other hand, some other amino acid residues in NIGT1 may interact with the “G” in GAATC. In the binding of GAATATTC, the K285, W240, S282 and Q285 residues of NIGT1 participate in the interaction with the AAT base pairs, whereas the L284 residue would thus be free (). This DNA-protein complex is stable without the interaction of the L284 residue with a base, because both subunits of the dimer participate in the interaction with DNA. It is likely that the accessibility of the L284 residue to DNA may be restricted in a dimer conformation and therefore that the dimer preferentially binds to the GAATATTC sequence. However, I cannot rule out the possibility that the dimer may also bind to GAATC using only one monomer subunit, because the interaction between NIGT1 and its GAATC recognition sequence is inhibited by the presence of a GAATATTC sequence in an in vitro binding experiment.Citation11
This hypothesis to explain the dual specificity of NIGT1 for DNA binding can also explain differences in the recognition sequences of GARP family proteins. GLK1 and GLK2 bind to CCAATC probably as monomers, because the GLK-binding sequence contains AATC and residues that interact with this site are conserved in both GLK1 and GLK2. In the case of PHR1, the S272, Q275, W230 and K276 residues in each subunit of a dimer likely interact with GNAT sites, which represent a half portion of the PHR1 binding site. Unexpectedly, it has been found that the residue corresponding to L231 of ARR10 and L284 of NIGT1 is conserved as L274 in PHR1. The L274 residue does not participate in the interaction with DNA but might be necessary to stabilize the protein structure due to its central location in the GARP domain.
In my hypothesis therefore, the GARP domain is a backbone for interactions with similar DNA sequences containing an AATC sequence, but amino acid substitutions will affect the interactions between the GARP domain and the sequences flanking the AATC sequence. The more important point in this regard is that dimer formation allows selective interactions with only some bases in the AATC motif to enable the formation of stable DNA-protein complexes. Hence, the dimer can bind to more a divergent range of DNA sequences. It is also worth noting that, if this hypothesis is correct, dimerization and dissociation of GARP family proteins would likely function as a molecular mechanism for altering target genes. As studies of these proteins are currently limited, and current evidence is therefore sparse, further characterization of the DNA binding of the GARP family proteins will be essential to test this hypothesis.
Acknowledgments
This work was supported in part by KAKENHI (22380043) from the Japan Society for the Promotion of Science (JSPS) and a Grant-in-Aid for Scientific Research on Innovative Areas (21114004) from the Ministry of Education, Culture, Sports, Science and Technology of Japan (MEXT).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Riechmann JL, Heard J, Martin G, Reuber L, Jiang C, Keddie J, et al. Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 2000; 290:2105 - 10; http://dx.doi.org/10.1126/science.290.5499.2105; PMID: 11118137
- Qu LJ, Zhu Y-X. Transcription factor families in Arabidopsis: major progress and outstanding issues for future research. Curr Opin Plant Biol 2006; 9:544 - 9; http://dx.doi.org/10.1016/j.pbi.2006.07.005; PMID: 16877030
- Hall LN, Rossini L, Cribb L, Langdale JA. GOLDEN 2: a novel transcriptional regulator of cellular differentiation in the maize leaf. Plant Cell 1998; 10:925 - 36; PMID: 9634581
- Imamura A, Hanaki N, Nakamura A, Suzuki T, Taniguchi M, Kiba T, et al. Compilation and characterization of Arabidopsis thaliana response regulators implicated in His-Asp phosphorelay signal transduction. Plant Cell Physiol 1999; 40:733 - 42; http://dx.doi.org/10.1093/oxfordjournals.pcp.a029600; PMID: 10501033
- Wykoff DD, Grossman AR, Weeks DP, Usuda H, Shimogawara K. Psr1, a nuclear localized protein that regulates phosphorus metabolism in Chlamydomonas.. Proc Natl Acad Sci U S A 1999; 96:15336 - 41; http://dx.doi.org/10.1073/pnas.96.26.15336; PMID: 10611385
- Rubio V, Linhares F, Solano R, Martín AC, Iglesias J, Leyva A, et al. A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev 2001; 15:2122 - 33; http://dx.doi.org/10.1101/gad.204401; PMID: 11511543
- Waters MT, Moylan EC, Langdale JA. GLK transcription factors regulate chloroplast development in a cell-autonomous manner. Plant J 2008; 56:432 - 44; http://dx.doi.org/10.1111/j.1365-313X.2008.03616.x; PMID: 18643989
- Waters MT, Wang P, Korkaric M, Capper RG, Saunders NJ, Langdale JA. GLK transcription factors coordinate expression of the photosynthetic apparatus in Arabidopsis. Plant Cell 2009; 21:1109 - 28; http://dx.doi.org/10.1105/tpc.108.065250; PMID: 19376934
- Onai K, Ishiura M. PHYTOCLOCK 1 encoding a novel GARP protein essential for the Arabidopsis circadian clock. Genes Cells 2005; 10:963 - 72; http://dx.doi.org/10.1111/j.1365-2443.2005.00892.x; PMID: 16164597
- Hosoda K, Imamura A, Katoh E, Hatta T, Tachiki M, Yamada H, et al. Molecular structure of the GARP family of plant Myb-related DNA binding motifs of the Arabidopsis response regulators. Plant Cell 2002; 14:2015 - 29; http://dx.doi.org/10.1105/tpc.002733; PMID: 12215502
- Sawaki N, Tsujimoto R, Shigyo M, Konishi M, Toki S, Fujiwara T, et al. A nitrate-inducible GARP family gene encodes an auto-repressible transcriptional repressor in rice. Plant Cell Physiol 2013; In press http://dx.doi.org/10.1093/pcp/pct007; PMID: 23324170