Abstract
Auxin, a plant hormone, plays crucial roles in diverse aspects of plant growth and development reacting to and integrating environmental stimuli. Indole-3-acetic acid (IAA) is the major plant auxin that is synthesized by members of the YUCCA (YUC) family of flavin monooxygenases that catalyse a rate-limiting step. Although the paths to IAA biosynthesis are characterized in Arabidopsis, little is known about the corresponding components in potato. Recently, we isolated eight putative StYUC (Solanum tuberosum YUCCA) genes and five putative tryptophan aminotransferase genes in comparison to those found in Arabidopsis.Citation1 The specific domains of YUC proteins were well conserved in all StYUC amino acid sequences. Transgenic potato (Solanum tuberosum cv. Jowon) overexpressing AtYUC6 showed high-auxin and enhanced drought tolerance phenotypes. The transgenic potatoes also exhibited reduced levels of ROS (reactive oxygen species) compared to control plants. We therefore propose that YUCCA and TAA families in potato would function in the auxin biosynthesis. The overexpression of AtYUC6 in potato establishes enhanced drought tolerance through regulated ROS homeostasis.
Plants produce various phytohormones, including auxins, gibberellins, cytokinins, ethylene, abscisic acid and brassinosteroids. Their synthesis is delicately regulated to orchestrate normal cell, organ and plant growth and development. In addition, the phytohormones co-adjust agonistically or antagonistically to demands originating from environmental cues. Among the phytohormones, auxin is essential as a regulator of growth and development, involved in diverse processes, such as cell division, expansion and differentiation, and also in lateral root formation, flowering, tropic responses, and senescence.Citation2-Citation5 Recent studies also provided evidence for the function of auxin in responses to environmental stresses, including drought, salinity and pathogen attack.Citation6-Citation8
Indole-3-acetic acid (IAA) is the main plant auxin synthesized by both tryptophan (Trp)-dependent and -independent pathways.Citation9 Although molecular components and physiological functions of the Trp-independent pathway are unknown, the Trp-dependent pathway is well defined as multiple pathways that proceed through four metabolic intermediates.Citation5 These can be divided into the indole-3-acetaldoxime (IAOx), indole-3-acetamide (IAM), tryptamine (TAM) and indole-3-pyruvic acid (IPA) pathways. Genetic and biochemical studies in Arabidopsis have shown the preponderance of the Trp-dependent pathway in de novo auxin biosynthesis. Evidently the Trp-dependent pathway is involved in embryogenesis, seedling growth, flower development, vascular patterning, while it affects other developmental processes as well.Citation10-Citation13
Significantly, the IPA pathway constitutes a simple two-step pathway in Arabidopsis.Citation14,Citation15 The first step is the conversion of tryptophan to indole-3-pyruvic acid (IPA) by a family of tryptophan aminotransferase of Arabidopsis (TAA). The TAA family consists of three closely related genes in Arabidopsis (TAA1, TAR1 and TAR2). Mutations of these genes resulted in partial auxin deficiency phenotypically revealed by altered responses to shade avoidance, ethylene and auxin transport inhibitors, respectively.Citation12,Citation13,Citation16The defects of taa mutants could be partially rescued by auxin supplemented to growth media.Citation12,Citation13 These results strongly suggest that the TAA family genes constitute essential components for auxin biosynthesis. This fact suggested detailed studies on functions of TAA family genes in crop plants. We focused Solanum tuberosum, potato, in an attempt to define and isolate putative TAA gene family members. Based on the potato genome database (solanaceae.plantbiology.msu.edu/pgsc_download.shtml), we isolated five putative TAA family genes, termed Solanum tuberosum TRYPTOPHAN AMINOTRANSFERASE RELATED1 to 5 (StTAR1 to 5). The StTARs showed approximately 50% deduced amino acid sequence identity with Arabidopsis TAA/TAR1/TAR2. Also present in the potato sequences are the characteristic alliinase C and aromatic aminotransferase domains of TAA proteins.Citation1
In addition, the rate-limiting second step in Trp-dependent auxin biosynthesis is conversion of IPA to IAA by members of the YUCCA family of flavin monooxygenases (FMOs). Previously, the involvement of YUCCA proteins in the TAM pathway, a different Trp-dependent auxin biosynthesis pathway was reported because of the ability of YUCCA proteins to use TAM as a substrate.Citation5 This view has been replaced by recent studies in Arabidopsis that revealed the function of YUCCA proteins in the IPA pathway with TAA/TARs.Citation14,Citation15 In Arabidopsis, the YUCCA family consists of 11 genes. Lines overexpressing the proteins and activation-tagged mutants of individual genes showed auxin overproduction phenotypes, such as elongated hypocotyl, epinastic cotyledons, curled-down rosette leaves and strong apical dominance.Citation8,Citation17-Citation20 However, single YUC gene mutations fail to show a particular phenotype, suggesting overlapping functions for members of the family. For example, a quadruple mutant line, including yuc1yuc4yuc10yuc11, showed the classical developmental defects of auxin deficiency.Citation10,Citation11YUC genes have been identified as auxin-related genes in petunia, rice, corn and tomato.Citation21-Citation25 We now isolated eight putative YUC genes (StYUCs) from potato with 50% to70% amino acid identity with their Arabidopsis counterpart YUCCA proteins and including canonical, conserved YUCCA sequence domains. In addition, YUCCA6 overexpressed in Arabidopsis and potato plants led to auxin overproduction and drought tolerance phenotypes.Citation1 We surmise that the curled leaf structure observed in YUCCA6-overexpressing plants may cause a decrease in transpiration that could confer or support drought tolerance, but direct evidence is yet to be provided.
Several phytohormones are involved in stress responses. Examples are the GA repressor DELLA in salt stress and SA repressor JAZ in pathogen infections.Citation26,Citation27 Networks of ROS signaling in the chloroplast and mitochondria play essential roles in response to abiotic stresses in plants.Citation28 In addition, the crosstalk between auxin regulatory networks and ROS regulates the integration of environmental stress signals.Citation29 Auxin and ROS homeostasis would be interconnected by a redox balance, which could control the environmental stresses. However, an effect for auxin has not yet been clearly identified. yuc6-1D showed lower accumulation of hydrogen peroxide, a reactive oxygen species (ROS), compared to wild-type plants, while other antioxidant genes are not enhanced by YUCCA6 overexpression. In plants subjected to environmental stresses, ROS and Ca2+ accumulate significantly in plant cells. Thus, low concentrations of ROS in yuc6-1D could be at the basis of the observed drought tolerance. Recently, YUCCA7 transcript was observed to increase under drought stress, but YUCCA7 overexpression failed to correlate with drought tolerance in an ABA dependent manner.Citation8 This could imply that drought tolerance by YUCCA6 overexpression may include post-transcriptional control or the involvement of an unknown ROS scavenging system induced by YUCCA6 overexpression to maintain the ROS homeostasis (). One recent observation could point in this direction as well. Disruption of thioredoxin and glutathione systems resulted in the inhibition of auxin transport displaying the inflorescence stem, pin-like phenotypes, indicating that auxin signaling has some interplay with thioredoxin and glutathione systems.Citation30,Citation31 In addition, it is reported that auxin and ABA interplay through the reduced auxin levels caused by mitochondrial ROS overexpression.Citation32 These may correlate with unknown functions of YUCCA6 in conferring drought tolerance. Further investigations into the stress signaling responses related to auxin’s action will be necessary. This will provide better understanding of the roles of YUCCAs in the cross-talks between auxin dynamics and drought stress responses of plants.
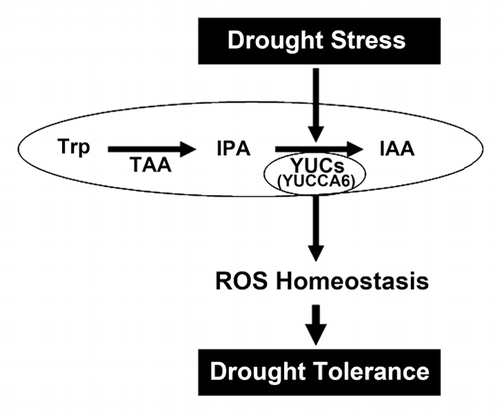
Figure 1. Model proposed for the function of YUCCA6 in drought stress. YUCCA6 protein is involved in auxin biosynthesis as a rate-limiting step converting IPA to IAA. It also functions in a drought stress signaling pathway through effects of ROS equilibrium in plants. Trp, tryptophan; TAA, tryptophan aminotransferase of Arabidopsis; IPA, indole-3-phyruvic acid; IAA, indole-3-acetic acid; ROS, reactive oxygen species.
Acknowledgments
We thank Dr Hans J. Bohnert for critical reading and insightful comments. This work was supported by grants from the World Class University Program (R32-10148) funded by the Ministry of Education, Science and Technology and Next-Generation BioGreen21 Program (SSAC, grant#: PJ009557), Rural Development Administration, Republic of Korea.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Kim JI, Baek D, Park HC, Chun HJ, Oh DH, Lee MK, et al. Overexpression of Arabidopsis YUCCA6 in potato results in high-auxin developmental phenotypes and enhanced resistance to water deficit. . Mol Plant 2013; 6:337 - 49; PMID: 22986790
- Ellis CM, Nagpal P, Young JC, Hagen G, Guilfoyle TJ, Reed JW. AUXIN RESPONSE FACTOR1 and AUXIN RESPONSE FACTOR2 regulate senescence and floral organ abscission in Arabidopsis thaliana.. Development 2005; 132:4563 - 74; http://dx.doi.org/10.1242/dev.02012; PMID: 16176952
- Davis PJ. The Plant Hormones: Their Nature, Occurrence, and Functions. (Kluwer, Dordrecht, The Netherlands). Plant Hormones 2010;1-15.
- Kim JI, Murphy AS, Baek D, Lee S-W, Yun D-J, Bressan RA, et al. YUCCA6 over-expression demonstrates auxin function in delaying leaf senescence in Arabidopsis thaliana.. J Exp Bot 2011; 62:3981 - 92; http://dx.doi.org/10.1093/jxb/err094; PMID: 21511905
- Rosquete MR, Barbez E, Kleine-Vehn J. Cellular auxin homeostasis: gatekeeping is housekeeping. Mol Plant 2012; 5:772 - 86; http://dx.doi.org/10.1093/mp/ssr109; PMID: 22199236
- Park C-M. Auxin homeostasis in plant stress adaptation response. Plant Signal Behav 2007; 2:306 - 7; http://dx.doi.org/10.4161/psb.2.4.4069; PMID: 19704630
- Ghanashyam C, Jain M. Role of auxin-responsive genes in biotic stress responses. Plant Signal Behav 2009; 4:846 - 8; http://dx.doi.org/10.4161/psb.4.9.9376; PMID: 19847104
- Lee M, Jung J-H, Han D-Y, Seo PJ, Park WJ, Park C-M. Activation of a flavin monooxygenase gene YUCCA7 enhances drought resistance in Arabidopsis.. Planta 2012; 235:923 - 38; http://dx.doi.org/10.1007/s00425-011-1552-3; PMID: 22109847
- Zhao Y. Auxin biosynthesis and its role in plant development. Annu Rev Plant Biol 2010; 61:49 - 64; http://dx.doi.org/10.1146/annurev-arplant-042809-112308; PMID: 20192736
- Cheng Y, Dai X, Zhao Y. Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis.. Genes Dev 2006; 20:1790 - 9; http://dx.doi.org/10.1101/gad.1415106; PMID: 16818609
- Cheng Y, Dai X, Zhao Y. Auxin synthesized by the YUCCA flavin monooxygenases is essential for embryogenesis and leaf formation in Arabidopsis.. Plant Cell 2007; 19:2430 - 9; http://dx.doi.org/10.1105/tpc.107.053009; PMID: 17704214
- Stepanova AN, Robertson-Hoyt J, Yun J, Benavente LM, Xie D-Y, Doležal K, et al. TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 2008; 133:177 - 91; http://dx.doi.org/10.1016/j.cell.2008.01.047; PMID: 18394997
- Tao Y, Ferrer JL, Ljung K, Pojer F, Hong F, Long JA, et al. Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 2008; 133:164 - 76; http://dx.doi.org/10.1016/j.cell.2008.01.049; PMID: 18394996
- Won C, Shen X, Mashiguchi K, Zheng Z, Dai X, Cheng Y, et al. Conversion of tryptophan to indole-3-acetic acid by TRYPTOPHAN AMINOTRANSFERASES OF ARABIDOPSIS and YUCCAs in Arabidopsis.. Proc Natl Acad Sci USA 2011; 108:18518 - 23; http://dx.doi.org/10.1073/pnas.1108436108; PMID: 22025721
- Mashiguchi K, Tanaka K, Sakai T, Sugawara S, Kawaide H, Natsume M, et al. The main auxin biosynthesis pathway in Arabidopsis.. Proc Natl Acad Sci USA 2011; 108:18512 - 7; http://dx.doi.org/10.1073/pnas.1108434108; PMID: 22025724
- Yamada M, Greenham K, Prigge MJ, Jensen PJ, Estelle M. The TRANSPORT INHIBITOR RESPONSE2 gene is required for auxin synthesis and diverse aspects of plant development. Plant Physiol 2009; 151:168 - 79; http://dx.doi.org/10.1104/pp.109.138859; PMID: 19625638
- Zhao Y, Christensen SK, Fankhauser C, Cashman JR, Cohen JD, Weigel D, et al. A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 2001; 291:306 - 9; http://dx.doi.org/10.1126/science.291.5502.306; PMID: 11209081
- Marsch-Martinez N, Greco R, Van Arkel G, Herrera-Estrella L, Pereira A. Activation tagging using the En-I maize transposon system in Arabidopsis.. Plant Physiol 2002; 129:1544 - 56; http://dx.doi.org/10.1104/pp.003327; PMID: 12177467
- Woodward C, Bemis SM, Hill EJ, Sawa S, Koshiba T, Torii KU. Interaction of auxin and ERECTA in elaborating Arabidopsis inflorescence architecture revealed by the activation tagging of a new member of the YUCCA family putative flavin monooxygenases. Plant Physiol 2005; 139:192 - 203; http://dx.doi.org/10.1104/pp.105.063495; PMID: 16126863
- Kim JI, Sharkhuu A, Jin JB, Li P, Jeong JC, Baek D, et al. yucca6, a dominant mutation in Arabidopsis, affects auxin accumulation and auxin-related phenotypes. Plant Physiol 2007; 145:722 - 35; http://dx.doi.org/10.1104/pp.107.104935; PMID: 17885085
- Tobeña-Santamaria R, Bliek M, Ljung K, Sandberg G, Mol JNM, Souer E, et al. FLOOZY of petunia is a flavin mono-oxygenase-like protein required for the specification of leaf and flower architecture. Genes Dev 2002; 16:753 - 63; http://dx.doi.org/10.1101/gad.219502; PMID: 11914280
- Woo YM, Park HJ, Su’udi M, Yang JI, Park JJ, Back K, et al. Constitutively wilted 1, a member of the rice YUCCA gene family, is required for maintaining water homeostasis and an appropriate root to shoot ratio. Plant Mol Biol 2007; 65:125 - 36; http://dx.doi.org/10.1007/s11103-007-9203-6; PMID: 17619151
- Yamamoto Y, Kamiya N, Morinaka Y, Matsuoka M, Sazuka T. Auxin biosynthesis by the YUCCA genes in rice. Plant Physiol 2007; 143:1362 - 71; http://dx.doi.org/10.1104/pp.106.091561; PMID: 17220367
- Expósito-Rodríguez M, Borges AA, Borges-Pérez AB, Hernández M, Pérez JA. Cloning and biochemical characterization of ToFZY, a tomato gene encoding a flavin monooxygenase involved in a tryptophan-dependent auxin biosynthesis pathway. J Plant Growth Regul 2007; 26:329 - 40; http://dx.doi.org/10.1007/s00344-007-9019-2
- Gallavotti A, Barazesh S, Malcomber S, Hall D, Jackson D, Schmidt RJ, et al. sparse inflorescence1 encodes a monocot-specific YUCCA-like gene required for vegetative and reproductive development in maize. Proc Natl Acad Sci USA 2008; 105:15196 - 201; http://dx.doi.org/10.1073/pnas.0805596105; PMID: 18799737
- Achard P, Cheng H, De Grauwe L, Decat J, Schoutteten H, Moritz T, et al. Integration of plant responses to environmentally activated phytohormonal signals. Science 2006; 311:91 - 4; http://dx.doi.org/10.1126/science.1118642; PMID: 16400150
- Van der Does D, Leon-Reyes A, Koornneef A, Van Verk MC, Rodenburg N, Pauwels L, et al. Salicylic acid suppresses jasmonic acid signaling downstream of SCFCOI1-JAZ by targeting GCC promoter motifs via transcription factor ORA59. Plant Cell 2013; 25:744 - 61; In press http://dx.doi.org/10.1105/tpc.112.108548; PMID: 23435661
- Suzuki N, Koussevitzky S, Mittler R, Miller G. ROS and redox signalling in the response of plants to abiotic stress. Plant Cell Environ 2012; 35:259 - 70; http://dx.doi.org/10.1111/j.1365-3040.2011.02336.x; PMID: 21486305
- Tognetti VB, Mühlenbock P, Van Breusegem F. Stress homeostasis - the redox and auxin perspective. Plant Cell Environ 2012; 35:321 - 33; http://dx.doi.org/10.1111/j.1365-3040.2011.02324.x; PMID: 21443606
- Bashandy T, Guilleminot J, Vernoux T, Caparros-Ruiz D, Ljung K, Meyer Y, et al. Interplay between the NADP-linked thioredoxin and glutathione systems in Arabidopsis auxin signaling. Plant Cell 2010; 22:376 - 91; http://dx.doi.org/10.1105/tpc.109.071225; PMID: 20164444
- Koprivova A, Mugford ST, Kopriva S. Arabidopsis root growth dependence on glutathione is linked to auxin transport. Plant Cell Rep 2010; 29:1157 - 67; http://dx.doi.org/10.1007/s00299-010-0902-0; PMID: 20669021
- He J, Duan Y, Hua D, Fan G, Wang L, Liu Y, et al. DEXH box RNA helicase-mediated mitochondrial reactive oxygen species production in Arabidopsis mediates crosstalk between abscisic acid and auxin signaling. Plant Cell 2012; 24:1815 - 33; http://dx.doi.org/10.1105/tpc.112.098707; PMID: 22652060