Abstract
Alternative splicing (AS) gives rise to multiple mRNA isoforms from the same gene, providing possibilities to regulate gene expression beyond the level of transcription. In a recent paper in Nucleic Acids Research we used a high resolution RT-PCR based panel to study changes in AS patterns in plants with altered levels of an hnRNP-like RNA-binding protein in Arabidopsis thaliana. Furthermore, we detected significant changes in AS patterns between different Arabidopsis ecotypes. Here we investigated how small changes in ambient temperature affect AS. We found significant changes in AS for 12 of 28 investigated events (43%) upon transfer of Arabidopsis plants from 20°C to 16°C and for 6 of the 28 investigated events (21%) upon transfer from 20°C to 24°C.
Primary transcripts or precursor mRNAs (pre-mRNAs) usually contain introns that are removed during splicing. By combinatorial use of alternative splice sites pre-mRNAs can produce multiple transcript isoforms, leading to protein variants with different domains.Citation1,Citation2 Furthermore, different transcript isoforms originating from the same pre-mRNA can have different fates in the cell due to the presence or absence of miRNA target sites or inclusion of premature termination codons (PTC), channeling the transcript into the nonsense-mediated decay (NMD) pathway.Citation3-Citation6 Because recent data show that more than 60% of transcripts in Arabidopsis are alternatively spliced under regular growth conditions, AS provides an important layer of regulation at the posttranscriptional level.Citation7 In particular, AS patterns of disease resistance genes have been shown to change upon pathogen infection.Citation8-Citation10 Furthermore, abiotic stresses such as cold or heat treatment elicit changes in AS patterns.Citation1,Citation11,Citation12 The use of AS sites is determined by RNA-binding proteins, predominantly SR (serine/arginine)-rich proteins and hnRNPs (heterogenous nuclear ribonucleoproteins) which help to recruit the spliceosome to the splice sites.Citation13-Citation15
Previously, we employed a high resolution AS panel based on RT-PCR with fluorescent primers to investigate the impact of the hnRNP-like RNA-binding protein AtGRP7 (Arabidopsis thaliana glycine-rich RNA-binding protein 7)Citation16,Citation17 on a suite of known AS events in Arabidopsis. For 59 out of 288 analyzed AS events (21%) the ratio between the main splice isoforms significantly changed by more than 5% (p < 0.05) in plants constitutively overexpressing this RNA-binding protein (AtGRP7-ox plants) compared with wild type plants.Citation18 Notably, we found ten AS events that showed a reciprocal change in the ratio of AS forms in plants with elevated levels of AtGRP7 compared with plants with reduced levels of AtGRP7. This suggested that AtGRP7 directly regulates these AS events. Indeed, RNA immunoprecipitation experiments revealed that the majority of these transcripts were bound in vivo by AtGRP7. In addition, mutation of a single conserved arginine that impairs the AtGRP7 RNA-binding capability abrogates the effect of AtGRP7 on AS of these events indicating that the effect of AtGRP7 on AS relies on RNA binding.Citation18,Citation19 Comparison with the analysis of plants with altered levels of SR proteins uncovered several events that are controlled by both, AtGRP7 and SR proteins.Citation20 Recently, the hnRNP-like polypyrimidine tract binding proteins (PTBs) also have been shown to globally impact AS in Arabidopsis. Around 450 AS events react to changes in PTB1 and PTB2 levels.Citation21 It is not yet known how many of the transcripts are directly bound by the PTBs.
In the course of monitoring AS in two independent transgenic AtGRP7-ox lines generated in two different ecotypes, we also compared a suite of AS events between the Col and the C24 ecotypes themselves. We found that around 30% of the AS events changed significantly (> 5%, p < 0.05) between the ecotypes. We did not find single nucleotide polymorphisms in the splice sites per se but in intronic or exonic sequences near the splice sites that may cause ecotype-specific usage of the AS sites in either ecotype.Citation18
Ratios of AS Isoforms are Altered in Response to Small Temperature Changes
Several studies have shown that exposure to temperature stresses influences AS patterns in plants.Citation1,Citation11,Citation22-Citation24 However, the impact of small changes in ambient temperature has not been addressed. To investigate how a temperature step as small as 4°C affects AS, Arabidopsis plants were grown at 20°C and subsequently transferred to either 16°C or 24°C for one day in three biological replicates. The corresponding cDNAs were analyzed on the RT-PCR based AS panel. Of the 59 AS events previously investigated in detailCitation18 we selected 28 AS events in transcripts encoding RNA processing factors, transcription factors and other predicted regulatory proteinsCitation20 ().
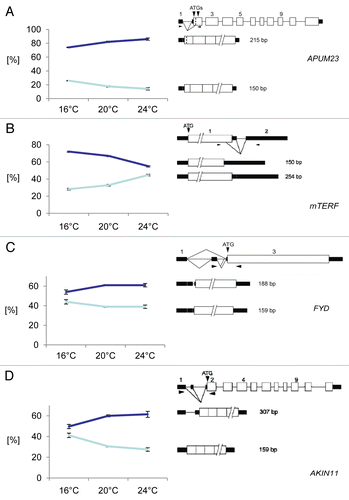
For 12 of 28 investigated AS events (43%), the ratio of the two splice isoforms changed more than 4% (p < 0.05) in wild type plants transferred to 16°C relative to control plants kept at 20°C (, ). For 6 of 28 investigated AS events (21%) the ratio showed a significant change in wild type plants transferred to 24°C relative to control plants kept at 20°C (, ). Four transcripts showed AS changes upon transfer from 20°C to both 16°C and 24°C (, ). For At1g72320 encoding APUM23,Citation25 a member of the PUMILIO family of RNA-binding proteins, alternative 3′ splice sites at the 5′ UTR intron lead to transcripts which either encode the authentic protein (according to gene models from TAIR) corresponding to a 215 nt PCR product or an N-terminally truncated protein corresponding to a 150 nt PCR product due to removal of part of exon2 including the authentic start codon (). Upon increasing the temperature an increase in the relative proportion of the 215 nt form corresponding to the authentic protein and a concomitant decrease in the 150 nt form is observed. For At2g3600 encoding a mitochondrial transcription termination factor-related protein (mTERF) the use of the authentic 5′ splice site of the intron in the 3′UTR leads to the mRNA (corresponding to a 254 nt PCR product) which gives rise to the authentic protein (according to gene models from TAIR) (). The use of an alternative 5′ splice site toward the end of the coding region gives an AS variant (corresponding to the 150 nt product) which would lead to a C-terminally truncated protein. With decreasing temperature there is a shift toward the form encoding the authentic protein (). The primers for FYD (At3g12570) cover an alternative 3′ splice site in intron2 in the 5′ UTR (). Upon decreasing temperature from 20°C to 16°C, a reduction of the large variant retaining part of intron2 is observed. This transcript isoform is stabilized in plants treated with cycloheximide, suggesting that it is an NMD substrate.Citation6 Finally, for AKIN11 encoding a catalytic subunit of Snf1-related protein kinase, primers detect the use of an alternative 5′ splice site in intron1 in the 5′ UTR. Upon decreasing temperature from 20°C to 16°C the proportion of the longer splice variant retaining part of the intron is reduced ().
Figure 1: Temperature dependence of AS events that change upon increasing and decreasing ambient temperature. (A) APUM23; (B) mTERF; (C) FYD; (D) AKIN11.The gene structure, the structure of the transcript isoforms around the AS event and the ratios of the two splice isoforms at each temperature are shown. On the left side of each panel, the percentage of each splice form +/− s.d. based on three biological replicates is indicated for each temperature. On the right side of each panel, the gene and transcript structures and the AS events are shown schematically. Exons are indicated by open boxes; UTRs, black rectangles; introns, thin lines; splicing events, diagonal lines. The arrowheads denote the approximate position of primers and the sizes of the PCR products from each splice isoform are indicated.
The observed changes in the ratios of AS isoforms probably are a consequence of a differential choice of AS sites in response to temperature. However, it is possible that such changes could occur indirectly as a consequence of altered steady-state abundance at the different temperatures potentially reflecting transcription or stability. Thus, the transcript levels were calculated relative to the reference transcript RPL12c. None of the affected transcripts showed a statistically significant difference in steady-state abundance of at least 2-fold when plants were transferred to 16°C or 24°C ().
In terms of stability of the alternatively spliced transcripts, some of the transcripts studied here are known to undergo NMDCitation6 but did not show consistent effects at different temperatures. For example, the transcript represented by the 215 bp product of APUM23 whose level decreases at 16°C and increases at 24°C is an NMD substrateCitation6 (); in contrast, the 190 bp product of At2g04790 (unknown protein), also an NMD substrate,Citation6 is decreased at both 16°C and 24°C (). Similarly, the level of the 159 bp product of AKIN11, another NMD substrate,Citation6 increased at 16°C while other NMD transcripts (e.g., APUM23, 259 bp ; and At2g04790, 190 bp) decreased at 16°C (, ). In addition, changes in AS seen in a previous study when plants are transferred to 4°C were not due to effects on NMD.Citation23 Thus, the observed changes are most likely to reflect alternative splicing.
Overall, our data show for the first time that changes in ambient temperature as small as 4°C can affect many AS events. Previously, larger temperature steps were shown to affect AS. For example, the transcripts encoding core components of the Arabidopsis circadian clock, CIRCADIAN ASSOCIATED1 and LATE ELONGATED HYPOCOTYL undergo AS upon exposure to 4°C.Citation22-Citation24 Because these AS events can lead to changes in transcript and protein levels they have been implicated in adjusting clock function to reduced ambient temperatures.Citation23,Citation24
Regulation of AS is determined by the relative abundance and activity of different splicing factors. This was illustrated in Arabidopsis by reciprocal changes in AS in plants with increased expression (overexpression lines) or no expression (mutant) of AtGRP7Citation18 and PTB proteins.Citation21 Here, four genes show reciprocal changes in AS in response to small increases and decreases in temperature suggesting temperature-dependent regulation. This most likely reflects altered composition or activity of splicing factorsCitation2 and it is notable that, for example, SR and hnRNP proteins themselves undergo AS in plants exposed to low or high temperatures, suggesting a global impact on downstream transcripts during temperature stress.Citation1,Citation14,Citation25,Citation26 This is further illustrated in a study on temperature-dependent flower induction, where an enrichment of RNA-processing related gene products was found upon transfer of Arabidopsis plants from 16°C to 25°C,Citation27,Citation28 and recently heat stress-induced AS was found to impact processing of a miRNA precursor located in the intron.Citation29 Changes in the use of splice sites could also involve temperature dependent secondary structure changes in the introns and adjacent exons. Techniques to assess the regulatory impact of RNA secondary structure at a global scale have recently been applied to Arabidopsis.Citation30
Conclusion and Perspective
We have shown here that changes in ambient temperature as small as 4°C can have a significant effect on AS events. Thus, plants may acclimate to changing environmental temperatures through adjusting the transcriptome by changes in AS patterns. This can involve altering levels of functional mRNAs able to encode protein by modulating the amount of AS giving rise to non-functional mRNAs, many of which are subject to NMD. Changes in AS can impact the proteome directly through translating AS isoforms into proteins with different domain structures that potentially fulfill temperature-specific functions. Finally, AS can have indirect consequences via interaction with the miRNA regulatory network, e.g., by temperature-conditional interference with processing of intronic miRNA precursors or altered miRNA target site availability.Citation4,Citation29 Overall, it will be important in future to identify all of the splicing factors involved in AS, what regulates their expression and activity and how they interact to determine the dynamic transcriptome and ultimately phenotype of cells and organisms.Citation31
Table 1: Details of the AS events analyzed.
Table 2: Transcripts with changes in AS patterns upon moderate temperature shifts.
Acknowledgments
This work was supported by an EMBO short-term fellowship to C.S., the German Research foundation (STA 653/2, SFB613) to D.S. and grants from the Biotechnology and Biological Sciences Research Council [BBSRC (BB/G024979/1)], European Research Area network (ERA-NET), Plant Genomics (Plant Alternative Splicing and Abiotic Stress) and the Scottish Government Rural and Environment Science and Analytical Services division (RESAS) to J.W.S.B.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Reddy AS. Alternative splicing of pre-messenger RNAs in plants in the genomic era. Annu Rev Plant Biol 2007; 58:267 - 94; http://dx.doi.org/10.1146/annurev.arplant.58.032806.103754; PMID: 17222076
- Syed NH, Kalyna M, Marquez Y, Barta A, Brown JWS. Alternative splicing in plants--coming of age. Trends Plant Sci 2012; 17:616 - 23; http://dx.doi.org/10.1016/j.tplants.2012.06.001; PMID: 22743067
- McGlincy NJ, Smith CW. Alternative splicing resulting in nonsense-mediated mRNA decay: what is the meaning of nonsense?. Trends Biochem Sci 2008; 33:385 - 93; http://dx.doi.org/10.1016/j.tibs.2008.06.001; PMID: 18621535
- Yang X, Zhang H, Li L. Alternative mRNA processing increases the complexity of microRNA-based gene regulation in Arabidopsis. Plant J 2012; 70:421 - 31; http://dx.doi.org/10.1111/j.1365-313X.2011.04882.x; PMID: 22247970
- Staiger D, Zecca L, Wieczorek Kirk DA, Apel K, Eckstein L. The circadian clock regulated RNA-binding protein AtGRP7 autoregulates its expression by influencing alternative splicing of its own pre-mRNA. Plant J 2003; 33:361 - 71; http://dx.doi.org/10.1046/j.1365-313X.2003.01629.x; PMID: 12535349
- Kalyna M, Simpson CG, Syed NH, Lewandowska D, Marquez Y, Kusenda B, et al. Alternative splicing and nonsense-mediated decay modulate expression of important regulatory genes in Arabidopsis. Nucleic Acids Res 2012; 40:2454 - 69; http://dx.doi.org/10.1093/nar/gkr932; PMID: 22127866
- Marquez Y, Brown JWS, Simpson CG, Barta A, Kalyna M. Transcriptome survey reveals increased complexity of the alternative splicing landscape in Arabidopsis. Genome Res 2012; 22:1184 - 95; http://dx.doi.org/10.1101/gr.134106.111; PMID: 22391557
- Dinesh-Kumar SP, Baker BJ. Alternatively spliced N resistance gene transcripts: their possible role in tobacco mosaic virus resistance. Proc Natl Acad Sci USA 2000; 97:1908 - 13; http://dx.doi.org/10.1073/pnas.020367497; PMID: 10660679
- Zhang X-C, Gassmann W. Alternative splicing and mRNA levels of the disease resistance gene RPS4 are induced during defense responses. Plant Physiol 2007; 145:1577 - 87; http://dx.doi.org/10.1104/pp.107.108720; PMID: 17951452
- Staiger D, Korneli C, Lummer M, Navarro L. Emerging role for RNA-based regulation in plant immunity. New Phytol 2013; 197:394 - 404; http://dx.doi.org/10.1111/nph.12022; PMID: 23163405
- Iida K, Seki M, Sakurai T, Satou M, Akiyama K, Toyoda T, et al. Genome-wide analysis of alternative pre-mRNA splicing in Arabidopsis thaliana based on full-length cDNA sequences. Nucleic Acids Res 2004; 32:5096 - 103; http://dx.doi.org/10.1093/nar/gkh845; PMID: 15452276
- Carvalho R, Feijao C, Duque P. On the physiological significance of alternative splicing events in higher plants. Protoplasma 2012; 249:1 - 12; PMID: 22160158
- Reddy ASN, Rogers MF, Richardson DN, Hamilton M, Ben-Hur A. Deciphering the plant splicing code: experimental and computational approaches for predicting alternative splicing and splicing regulatory elements. Front Plant Sci 2012; 3:18; http://dx.doi.org/10.3389/fpls.2012.00018; PMID: 22645572
- Barta A, Kalyna M, Reddy AS. Implementing a rational and consistent nomenclature for serine/arginine-rich protein splicing factors (SR proteins) in plants. Plant Cell 2010; 22:2926 - 9; http://dx.doi.org/10.1105/tpc.110.078352; PMID: 20884799
- Wachter A, Rühl C, Stauffer E. The role of polypyrimidine tract-binding proteins and other hnRNP proteins in plant splicing regulation. Front Plant Sci 2012; 3:81; http://dx.doi.org/10.3389/fpls.2012.00081; PMID: 22639666
- Heintzen C, Nater M, Apel K, Staiger D. AtGRP7, a nuclear RNA-binding protein as a component of a circadian-regulated negative feedback loop in Arabidopsis thaliana.. Proc Natl Acad Sci USA 1997; 94:8515 - 20; http://dx.doi.org/10.1073/pnas.94.16.8515; PMID: 9238008
- Heintzen C, Melzer S, Fischer R, Kappeler S, Apel K, Staiger D. A light- and temperature-entrained circadian clock controls expression of transcripts encoding nuclear proteins with homology to RNA-binding proteins in meristematic tissue. Plant J 1994; 5:799 - 813; http://dx.doi.org/10.1046/j.1365-313X.1994.5060799.x; PMID: 8054987
- Streitner C, Köster T, Simpson CG, Shaw P, Danisman S, Brown JWS, et al. An hnRNP-like RNA-binding protein affects alternative splicing by in vivo interaction with transcripts in Arabidopsis thaliana. Nucleic Acids Res 2012; 40:11240 - 55; http://dx.doi.org/10.1093/nar/gks873; PMID: 23042250
- Schöning JC, Streitner C, Meyer IM, Gao Y, Staiger D. Reciprocal regulation of glycine-rich RNA-binding proteins via an interlocked feedback loop coupling alternative splicing to nonsense-mediated decay in Arabidopsis. Nucleic Acids Res 2008; 36:6977 - 87; http://dx.doi.org/10.1093/nar/gkn847; PMID: 18987006
- Simpson CG, Fuller J, Maronova M, Kalyna M, Davidson D, McNicol J, et al. Monitoring changes in alternative precursor messenger RNA splicing in multiple gene transcripts. Plant J 2008; 53:1035 - 48; http://dx.doi.org/10.1111/j.1365-313X.2007.03392.x; PMID: 18088312
- Rühl C, Stauffer E, Kahles A, Wagner G, Drechsel G, Rätsch G, et al. Polypyrimidine tract binding protein homologs from Arabidopsis are key regulators of alternative splicing with implications in fundamental developmental processes. Plant Cell 2012; 24:4360 - 75; http://dx.doi.org/10.1105/tpc.112.103622; PMID: 23192226
- Filichkin SA, Priest HD, Givan SA, Shen R, Bryant DW, Fox SE, et al. Genome-wide mapping of alternative splicing in Arabidopsis thaliana.. Genome Res 2010; 20:45 - 58; http://dx.doi.org/10.1101/gr.093302.109; PMID: 19858364
- James AB, Syed NH, Bordage S, Marshall J, Nimmo GA, Jenkins GI, et al. Alternative splicing mediates responses of the Arabidopsis circadian clock to temperature changes. Plant Cell 2012; 24:961 - 81; http://dx.doi.org/10.1105/tpc.111.093948; PMID: 22408072
- James AB, Syed NH, Brown JW, Nimmo HG. Thermoplasticity in the plant circadian clock: how plants tell the time-perature. Plant Signal Behav 2012; 7:1219 - 23; http://dx.doi.org/10.4161/psb.21491; PMID: 22902701
- Tanabe N, Yoshimura K, Kimura A, Yabuta Y, Shigeoka S. Differential expression of alternatively spliced mRNAs of Arabidopsis SR protein homologs, atSR30 and atSR45a, in response to environmental stress. Plant Cell Physiol 2007; 48:1036 - 49; http://dx.doi.org/10.1093/pcp/pcm069; PMID: 17556373
- Lazar G, Goodman HM. The Arabidopsis splicing factor SR1 is regulated by alternative splicing. Plant Mol Biol 2000; 42:571 - 81; http://dx.doi.org/10.1023/A:1006394207479; PMID: 10809003
- Balasubramanian S, Weigel D. Temperature Induced Flowering in Arabidopsis thaliana. Plant Signal Behav 2006; 1:227 - 8; http://dx.doi.org/10.4161/psb.1.5.3452; PMID: 19704664
- Balasubramanian S, Sureshkumar S, Lempe J, Weigel D. Potent induction of Arabidopsis thaliana flowering by elevated growth temperature. PLoS Genet 2006; 2:e106; http://dx.doi.org/10.1371/journal.pgen.0020106; PMID: 16839183
- Yan K, Liu P, Wu C-A, Yang G-D, Xu R, Guo Q-H, et al. Stress-induced alternative splicing provides a mechanism for the regulation of microRNA processing in Arabidopsis thaliana. Mol Cell 2012; 48:521 - 31; http://dx.doi.org/10.1016/j.molcel.2012.08.032; PMID: 23063528
- Li F, Zheng Q, Vandivier LE, Willmann MR, Chen Y, Gregory BD. Regulatory impact of RNA secondary structure across the Arabidopsis transcriptome. Plant Cell 2012; 24:4346 - 59; http://dx.doi.org/10.1105/tpc.112.104232; PMID: 23150631
- Duque P. A role for SR proteins in plant stress responses. Plant Signal Behav 2011; 6:49 - 54; http://dx.doi.org/10.4161/psb.6.1.14063; PMID: 21258207