Abstract
Salt impairs cellular morphology and photosynthetic pigment accumulation in the cyanobacterium Fremyella diplosiphon. Recent findings indicated that the impact of salt on cellular morphology was attributable to salt-associated effects on osmotic regulation, as the impact on morphology was reversible when cells were treated with an osmoticum in the presence of salt. The impact of salt on photosynthetic pigment accumulation was associated with ionic effects of salt on the cells, as pigment levels remained low when salt-treated cells were incubated together with an osmoticum or an antioxidant, the latter to mitigate the impact of a salt-associated accumulation of reactive oxygen species. Here, we provide evidence that the transcripts for genes encoding the phycobiliproteins are not reduced in the presence of salt. These results suggest that the negative impact of salt-mediated changes on pigment accumulation occurs post-transcriptionally. A greater understanding of the mechanisms which impact growth of strains such as F. diplosiphon, which harbor pigments that allow low-light and shade-tolerated growth, may facilitate the development or adaptation of such strains as useful for remediation of salt-impacted soils or biofuel production.
Introduction
Increased salinity of water and soils negatively impacts the productivity of photosynthetic organisms.Citation1 Salinization is promoted by a number of factors, including human use of agricultural fertilizers,Citation2 road saltsCitation3 and other environmental factors, including drought and global temperature shifts.Citation4 As interest develops in using non-arable or brackish environments for growing bioenergy strains for the potential production of biofuels, it will be advantageous to use strains that can grow in a wide range of habitats, including those that are tolerant to low light and shade. Cyanobacteria and algae are becoming increasingly viewed as viable sources for producing biofuels for reasons that include their suitability for growth in saline-rich environments.Citation5 Using cyanobacterial or algal strains that also support growth in light-limited environments, including those strains that produce pigments such as phycoerythrin (PE) that allow growth in low light and shaded habitats,Citation6 would also likely increase their biofuel production capacity. Additionally, there has been interest in the use of salt-tolerant cyanobacterial strains for the remediation of salt-impacted soils.Citation7-Citation10
We decided to investigate the impacts of salinity on the growth and development of the freshwater cyanobacterium Fremyella diplosiphon, for which many studies have been conducted regarding the ability of the organism to adapt its growth dynamically to a range of conditions, including different colors and intensity of light.Citation11-Citation17 In recent experiments, we observed that salt stress alters growth, pigmentation and cellular morphology in F. diplosiphon.Citation18 In response to salt stress under both green light (GL) and red light (RL) conditions, photosynthetic pigment levels are reduced significantly and the morphology of cells are enlarged, indicative of stress.Citation18 Notably, an increased cell size in the presence of salt has been documented for other cyanobacteria.Citation19,Citation20 The salt-induced defect in morphology in F. diplosiphon is reversed to normal GL- and RL-specific cellular morphology when the osmoticum glycine betaine (GB) was added to the growth medium in the presence of salt.Citation18 However, GB was unable to restore the growth and pigmentation phenotypes to a level similar to that of control samples lacking salt, suggesting that morphological defects are induced by reversible osmotic effects of salt on cells, whereas pigmentation defects are caused by irreversible ionic toxicity caused by salt.Citation18
The impact of GB on salt-induced restoration of morphology in F. diplosiophon is attributed to cellular uptake of GB and the resultant restoration of osmotic pressure in the cell as a GB transport system is recognizable in the genome of F. diplosiphon (GenBank KC788749) and the genes are expressed as determined by RNA sequencing analyses.Citation18 The accumulation of phycobiliproteins (PBPs) and chlorophyll a (chla) were negatively impacted in F. diplosiphon in the presence of salt and this response was attributed to ionic imbalances as GB treatment or treatment with ascorbic acid (AA) to reverse oxidative stress did not restore pigment levels.Citation18 Here, we report on our complementary investigation which suggests that the impact of salt on PBP accumulation likely occurs at the post-transcriptional level.
Results
Salt does not reduce PBP gene transcript accumulation in F. diplosiphon
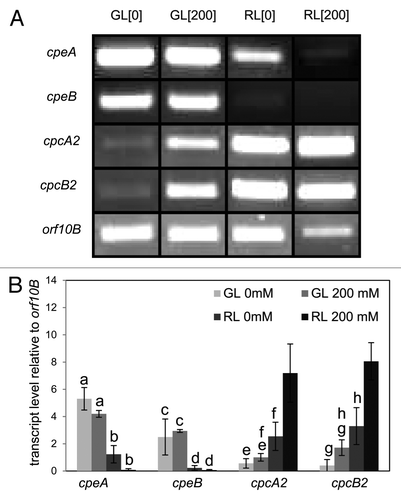
Once it became clear that GB could reverse the effect of salt on cellular morphology but had no major effect on restoring PBP concentrations,Citation18 we further investigated the effect of salt on transcript levels of genes encoding the α and β-subunits of PE (i.e., cpeBA) and inducible PC (i.e., cpcB2A2) under GL and RL using RT-PCR. The presence of salt did not significantly reduce transcript levels of genes encoding α and β-subunits of PE under GL (). However, under RL these cpeB and cpeA transcripts were low in the absence of salt as expected,Citation21,Citation22 and were not detected in the presence of salt (). Likewise, salt did not impair accumulation of the transcripts of both cpcB2 and cpcA2 under RL, compared to control samples lacking salt ().
Figure 1. Accumulation of phycobiliprotein gene transcripts in cells grown with or without sodium chloride (NaCl) salt under green light (GL) or red light (RL). RT-PCR analyses of the expression of cpeA, cpeB, cpcA2 and cpcB2 in Fremyella diplosiphon grown with (200 mM) or without (0 mM) NaCl under GL or RL. The transcript level of the orf10B gene was used as an internal control for each sample. (A) Representative agarose gel images and (B) average transcript levels relative to orf10B (± SD) calculated using densitometry measurements of three independent biological replicates. Identical letters over bars represent a homogeneous mean group (p > 0.05) within the bars for a single gene.
Discussion
In F. diplosiphon, a reduction in PC and PE content was found to be regulated at a post-transcriptional or translational level because salt addition to growth media did not impose any negative effect on accumulation of transcripts for genes encoding PC and PE monomers. Further investigation is required to understand how PC and PE are regulated post-transcriptionally or at the protein level in the presence of salt and how osmotic stress specifically may affect the cytoskeleton to alter cellular morphology under these conditions in F. diplosiphon.
Impacts of salt on growth and morphology of a range of other cyanobacteria and bacteria have been reported. The growth of the halotolerant cyanobacterium Aphanothece halophytica was reduced and bigger cell size was observed in the presence of > 0.5 M salt in the growth medium.Citation20 Growth was also reduced and a bigger cell size observed for the diazotrophic cyanobacterium Anabaena cylindrica.Citation19 Gram-negative, sulfate-reducing bacterium Desulfovibrio vulgaris exhibited elongated cell size by 5-fold in the presence of 250 mM NaCl.Citation23 However, the addition of GB was found to efficiently alleviate the morphological defects in this bacterium,Citation23 similar to what we observed for F. diplosiphon.Citation18 Reduced growth and larger cell shape with shorter interpeptide bridges in peptidoglycan were induced by 2.5 M NaCl in the Gram-positive bacterium Staphylococcus aureus; however, the presence of GB in the growth medium successfully reverted back to normal cell size and interpeptide peptidoglycon structure similar to control cells grown in the absence of salt.Citation24 These researchers concluded that high ionic strength is responsible for increased cell volume and altered shape.Citation24 The high osmolarity caused by NaCl or KCl inhibited cell division and resulted in the larger shape of Escherichia coli; however, addition of GB was found to restore DNA synthesis, cell division and normal cellular morphology.Citation25 All of these results, together with our prior recognition of the ability of salt to alter F. diplosiphon cell shape in a manner that can be reversed by GB,Citation18 suggest specific salt-associated disruptions in ionic strength or osmolarity that impair apposite regulation of cellular morphology. However, the mechanism is distinct from salt-mediated reduction in photosynthetic pigmentation and/or growth that is not GB-reversible, nor mediated by transcriptional downregulation of the phycobiliprotein genes in F. diplosiphon, the latter reported here.
Materials and Methods
Experimental organism and growth conditions
Wild-type pigmentation strain SF33Citation26 of F. diplosiphon was used in this study. Cells were grown in autoclaved BG-11 medium (Fluka) containing 10 mM HEPES (hereafter BG-11/HEPES) at pH 8.0 with or without 200 mM sodium chloride salt (NaCl) under continuous white fluorescent light (WL,:15 μmol m−2 s−1). WL-grown cultures in exponential phase were diluted to an initial OD750 of 0.2 and transferred to either GL or RL at: 15 μmol m−2 s−1 at 28°C with continuous shaking at ~175 rpm. GL and RL sources were those reported earlier.Citation13
Total RNA extraction and RT-PCR analysis
Once the OD750 reached more than 0.7 for all treatments, cultures were adjusted to an OD750 of ~0.7. RNA extraction was performed using Trizol reagent (1 ml) after overnight growth, as previously described.Citation27 Total RNA was treated to remove contaminating genomic DNA using a TURBO DNA-free kit (Ambion) according to the manufacturer’s instructions for rigorous DNase treatment in a 100 µl reaction volume. Following DNase treatment, a second Trizol extraction was performed to improve the quality of total RNA. Trizol (200 µl) and chloroform (40 µl) were added to the DNase-treated samples and incubated for 3 min after short vortexing. After centrifugation at 13,000 × g for 15 min at 4°C, the top colorless aqueous phase (200 µl) was transferred into a new Eppendorf tubes, followed by the addition of isopropanol (168 µl), with brief vortexing. After incubation for 15 min at room temperature, samples were centrifuged at 13,000 × g for 8 min and supernatant was discarded. Thereafter, the pellet was washed once with 1 ml of 70% ethanol and dried for 5 min at room temperature to remove the residual ethanol. Finally, the pellet was dissolved in RNase-free water and the quality and quantity of RNA determined spectrophotometrically by the A260/A280 ratio and assessed using agarose gel electrophoresis. Total RNA (0.5 µg) was reverse transcribed using random primers in a 20 µl reaction mixture using a Promega reverse transcription (RT) kit. No-RT reactions were also performed for all treatments where reverse transcriptase enzyme was not added to the RT reaction mixture. After RT, 180 µl of H2O was added to the mixture and 2 µl of it was used in conventional RT-PCR using GoTaq Green master mix (Promega) with primer sets designed for cpeA, cpeB, cpcA2, cpcB2 and internal control gene orf10B (for primer sequences see ). Orf10B was used as an internal control as this region was shown to be constantly expressed under GL and RL in a microarray study in F. diplosiphon.Citation28 PCR was performed using the following parameters: denaturation at 95°C for 1 min, then 30 (cpeA, cpeB, cpcA2, cpcB2) or 37 (orf10B) cycles of 95°C for 30 sec, annealing at the indicated temperature () for 30 sec and extension at 68°C for 30 sec, followed by an additional cycle of 68°C for 10 min, ending with a hold at 4°C. The PCR products were analyzed on a 0.8% agarose gel and imaged using Quantity One 1-D Software (Bio-Rad).
Table 1. Sequence of primers used in this study
| Abbreviations: | ||
| AP | = | allophycocyanin |
| chla | = | chlorophyll a |
| GL | = | green light |
| PBP | = | phycobiliprotein |
| PBS | = | phycobilisomes |
| PC | = | phycocyanin |
| PE | = | phycoerythrin |
| RL | = | red light |
| ROS | = | reactive oxygen species |
| WT | = | wild type |
Acknowledgments
This research was supported by the US Department of Energy (Chemical Sciences, Geosciences and Biosciences Division, Office of Basic Energy Sciences, Office of Science, grant no. DE-FG02-91ER20021 to B.L.M.). Support for Shailendra P. Singh was from a CAREER award from the National Science Foundation (grant no. MCB-0643516 to B.L.M.). We thank Britney Robinson, for whom funding was from the National Science Foundation (grant no. MCB-0919100 to B.L.M.) and Haley Miller, for whom funding was provided by the Professorial Assistantship Program of the Honors College at Michigan State University, for technical assistance. We also thank Jin Chen and Pingsha Hu for assistance with analyzing RNA sequencing data.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Epstein E, Bloom AJ. Mineral nutrition of plants: principles and perspectives. New York: John Wiley and Sons, 2005.
- Williams WD. Salinization of rivers and streams: an important environmental hazard. Ambio 1987; 16:180 - 5
- Kaushal SS, Groffman PM, Likens GE, Belt KT, Stack WP, Kelly VR, et al. Increased salinization of fresh water in the northeastern United States. Proc Natl Acad Sci USA 2005; 102:13517 - 20; http://dx.doi.org/10.1073/pnas.0506414102; PMID: 16157871
- Cañedo-Argüelles M, Kefford BJ, Piscart C, Prat N, Schäfer RB, Schulz CJ. Salinisation of rivers: an urgent ecological issue. Environ Pollut 2013; 173:157 - 67; http://dx.doi.org/10.1016/j.envpol.2012.10.011; PMID: 23202646
- Parmar A, Singh NK, Pandey A, Gnansounou E, Madamwar D. Cyanobacteria and microalgae: a positive prospect for biofuels. Bioresour Technol 2011; 102:10163 - 72; http://dx.doi.org/10.1016/j.biortech.2011.08.030; PMID: 21924898
- Dubinsky Z, Stambler N. Photoacclimation processes in phytoplankton: mechanisms, consequences, and applications. Aquat Microb Ecol 2009; 56:163 - 76; http://dx.doi.org/10.3354/ame01345
- Kaushik BD. Reclamative potential of cyanobacteria in salt-affected soils. Phykos 1989; 28:101 - 8
- Prasanna R, Jaiswal P, Kaushik BD. Cyanobacteria as potential options for environmental sustainability - promises and challenges. Indian J Microbiol 2008; 48:89 - 94; http://dx.doi.org/10.1007/s12088-008-0009-2; PMID: 23100703
- Subhashini D, Kaushik BD. Amelioration of sodic soils with blue-green algae. Aust J Soil Res 1981; 19:361 - 6; http://dx.doi.org/10.1071/SR9810361
- Rao DLN, Burns RG. The influence of blue-green algae on the biological amelioration of alkali soils. Biol Fertil Soils 1991; 11:306 - 12; http://dx.doi.org/10.1007/BF00335853
- Singh SP, Montgomery BL. Temporal responses of wild-type pigmentation and RcaE-deficient strains of Fremyella diplosiphon during light transitions. Commun Integr Biol 2011; b 4:503 - 10; PMID: 22046449
- Bennett A, Bogorad L. Complementary chromatic adaptation in a filamentous blue-green alga. J Cell Biol 1973; 58:419 - 35; http://dx.doi.org/10.1083/jcb.58.2.419; PMID: 4199659
- Bordowitz JR, Montgomery BL. Photoregulation of cellular morphology during complementary chromatic adaptation requires sensor-kinase-class protein RcaE in Fremyella diplosiphon.. J Bacteriol 2008; 190:4069 - 74; http://dx.doi.org/10.1128/JB.00018-08; PMID: 18390655
- Campbell D. Complementary chromatic adaptation alters photosynthetic strategies in the cyanobacterium Calothrix.. Microbiology 1996; 142:1255 - 63; http://dx.doi.org/10.1099/13500872-142-5-1255
- Kehoe DM, Grossman AR. Similarity of a chromatic adaptation sensor to phytochrome and ethylene receptors. Science 1996; 273:1409 - 12; http://dx.doi.org/10.1126/science.273.5280.1409; PMID: 8703080
- Pattanaik B, Whitaker MJ, Montgomery BL. Light quantity affects the regulation of cell shape in Fremyella diplosiphon.. Front Microbiol 2012; 3:170; http://dx.doi.org/10.3389/fmicb.2012.00170; PMID: 22586424
- Singh SP, Miller HL, Montgomery BL. . Temporal dynamics of changes in reactive oxygen species (ROS) levels and cellular morphology are coordinated during complementary chromatic adaptation in Fremyella diplosiphon.
- Singh SP, Montgomery BL. Salinity impacts photosynthetic pigmentation and cellular morphology changes by distinct mechanisms in Fremyella diplosiphon.. Biochem Biophys Res Commun 2013; 433:84 - 9; http://dx.doi.org/10.1016/j.bbrc.2013.02.060; PMID: 23454384
- Bhadauriya P, Gupta R, Singh S, Bisen PS. Physiological and biochemical alterations in a diazotrophic cyanobacterium Anabaena cylindrica under NaCl stress. Curr Microbiol 2007; 55:334 - 8; http://dx.doi.org/10.1007/s00284-007-0191-1; PMID: 17849161
- Laloknam S, Tanaka K, Buaboocha T, Waditee R, Incharoensakdi A, Hibino T, et al. Halotolerant cyanobacterium Aphanothece halophytica contains a betaine transporter active at alkaline pH and high salinity. Appl Environ Microbiol 2006; 72:6018 - 26; http://dx.doi.org/10.1128/AEM.00733-06; PMID: 16957224
- Oelmüller R, Conley PB, Federspiel NA, Briggs WR, Grossman AR. Changes in accumulation and synthesis of transcripts encoding phycobilisome components during acclimation of Fremyella diplosiphon to different light qualities. Plant Physiol 1988; 88:1077 - 83; http://dx.doi.org/10.1104/pp.88.4.1077; PMID: 16666425
- Oelmüller R, Grossman AR, Briggs WR. Photoreversibility of the effect of red and green light pulses on the accumulation in darkness of mRNAs coding for phycocyanin and phycoerythrin in Fremyella diplosiphon.. Plant Physiol 1988; 88:1084 - 91; http://dx.doi.org/10.1104/pp.88.4.1084; PMID: 16666426
- Mukhopadhyay A, He Z, Alm EJ, Arkin AP, Baidoo EE, Borglin SC, et al. Salt stress in Desulfovibrio vulgaris Hildenborough: an integrated genomics approach. J Bacteriol 2006; 188:4068 - 78; http://dx.doi.org/10.1128/JB.01921-05; PMID: 16707698
- Vijaranakul U, Nadakavukaren MJ, de Jonge BL, Wilkinson BJ, Jayaswal RK. Increased cell size and shortened peptidoglycan interpeptide bridge of NaCl-stressed Staphylococcus aureus and their reversal by glycine betaine. J Bacteriol 1995; 177:5116 - 21; PMID: 7665491
- Meury J. Glycine betaine reverses the effects of osmotic stress on DNA replication and cellular division in Escherichia coli.. Arch Microbiol 1988; 149:232 - 9; http://dx.doi.org/10.1007/BF00422010; PMID: 3284505
- Cobley JG, Zerweck E, Reyes R, Mody A, Seludo-Unson JR, Jaeger H, et al. Construction of shuttle plasmids which can be efficiently mobilized from Escherichia coli into the chromatically adapting cyanobacterium, Fremyella diplosiphon.. Plasmid 1993; 30:90 - 105; http://dx.doi.org/10.1006/plas.1993.1037; PMID: 8234495
- Seib LO, Kehoe DM. A turquoise mutant genetically separates expression of genes encoding phycoerythrin and its associated linker peptides. J Bacteriol 2002; 184:962 - 70; http://dx.doi.org/10.1128/jb.184.4.962-970.2002; PMID: 11807056
- Stowe-Evans EL, Ford J, Kehoe DM. Genomic DNA microarray analysis: identification of new genes regulated by light color in the cyanobacterium Fremyella diplosiphon.. J Bacteriol 2004; 186:4338 - 49; http://dx.doi.org/10.1128/JB.186.13.4338-4349.2004; PMID: 15205436