Abstract
The Arabidopsis genome encodes 10 Homeodomain-Leucine Zipper (HD-Zip) II transcription factors that can be subdivided into 4 clades (α–δ). All the γ (ARABIDOPSIS THALIANA HOMEOBOX 2 [ATHB2], HOMEOBOX ARABIDOPSIS THALIANA 1 [HAT1], HAT2) and δ (HAT3, ATHB4) genes are regulated by light quality changes (Low Red [R]/Far-Red [FR]) that induce the shade avoidance response in most of the angiosperms. HD-Zip IIγ and HD-Zip IIδ transcription factors function as positive regulators of shade avoidance, and there is evidence that at least ATHB2 is directly positively regulated by the basic Helix-Loop-Helix (bHLH) proteins PHYTOCHROME INTERACTING FACTOR 4 (PIF4) and PIF5. Recent evidence demonstrate that, in addition to their function in shade avoidance, HD-Zip IIγ and HD-Zip IIδ proteins play an essential role in plant development from embryogenesis onwards in a white light environment.
The HD-Zip class of proteins has been identified only in plants, and is characterized by a homeodomain linked to a leucine zipper motif.Citation1 Several Arabidopsis HD-Zip genes have been isolated in the 90s and, based on sequence homology in the HD-Zip domain, subdivided in 4 families, HD-Zip I–IV.Citation2 Determination of the Arabidopsis genome sequence has shown that it encodes for 48 HD-Zip proteins all members of the 4 families previously recognized.Citation2-Citation7 HD-Zip genes have been identified in a variety of land plants.Citation8
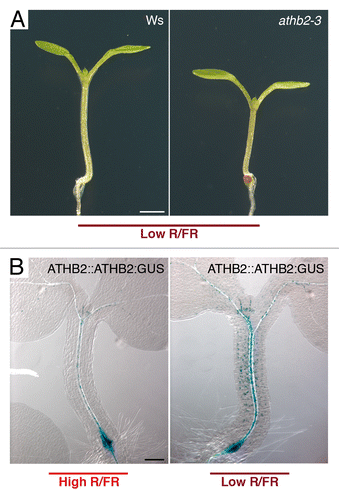
The HD-Zip II family comprises 10 members including ARABIDOPSIS THALIANA HOMOMEOBOX 2 (ATHB2Citation1; also known as HOMEOBOX ARABIDOPSIS THALIANA 4 [HAT4]Citation9), the first gene shown to be rapidly induced by changes in the Red (R) to Far-Red (FR) light ratio that promote the shade avoidance response in most angiosperms.Citation10,Citation11 It has been demonstrated that phytochromes B, D and E are all required for the regulation of ATHB2 by light quality changes,Citation11-Citation13 and evidence exist that this transcription factor gene is a direct target of the basic Helix-Loop-Helix (bHLH) transcription factors PHYTOCHROME INTERACTING FACTOR 4 (PIF4) and PIF5.Citation14 It has been proposed that ATHB2 functions as a positive regulator of shade avoidance,Citation15 and indeed loss-of-function athb2 mutants display diminished hypocotyl elongation in Low R/FR light with respect to wild-type seedlings (). Low R/FR light rapidly induces ATHB2:GUS expression in all cell layers of the elongating portion of the hypocotyl, thus suggesting that ATHB2 acts, at least in part, in this organ to control shade avoidance (). Links between ATHB2 and auxin, known to play a central role in shade avoidance,Citation14-Citation23 have been established.Citation15-Citation17 However, how ATHB2 promotes auxin response and/or transport in Low R/FR remains to be investigated.
Figure 1.ATHB2 is rapidly induced by low R/FR and required for shade-induced elongation. A) Wild-type (Ws) and athb2–328 (Ws) seedlings grown for 4 d in a light (L)/dark (D) cycle (16/8 h) in high R/FR, and then transferred to low R/FR under the same L/D regimen for 6 d (Low R/FR). No significant difference was observed in hypocotyl length in athb2-3 seedlings grown for 4 d in High R/FR relative to wild type (n ≥ 20). By contrast, athb2-3 seedlings elongate significantly less than wild type in Low R/FR (Ws:3.282 ± 0.078 mm; athb2-3:2.594 ± 0.101; n ≥ 30; P < 0.01). Bar: 1 mm. B) Histochemical localization of GUS activity in ATHB2::ATHB2:GUSCitation28 seedlings grown for 4 d in a L/D cycle (16/8 h) in high R/FR (High R/FR), and then exposed to low R/FR under the same L/D regimen for 2 h (Low R/FR). Bar: 0.1 mm. Growth conditions, light settings and GUS staining procedure were as previously reported.Citation19
Phylogeny reconstruction has indicated that the HD-Zip II genes can be grouped into 4 clades (α–δ).Citation7 All the γ (ATHB2, HAT1, HAT2) and δ genes (ATHB4, HAT3) have been shown to be induced by Low R/FR light, and kinetics of Low R/FR induction and reversibility by High R/FR have lead to the suggestion that HAT1, HAT3 and ATHB4, as ATHB2, are regulated by the phytochrome(s).Citation7 In addition, it has been shown that overexpression of ATHB2, HAT1, HAT2, initially identified as an auxin-inducible gene,Citation24 HAT3 and ATHB4 results in phenotypes resembling those of wild-type plants grown in Low R/FR light, further highlighting functional redundancy of HD-Zip IIγ and HD-Zip IIδ proteins in shade avoidance.Citation7,Citation15,Citation24-Citation27
Intriguingly, it has recently emerged that besides their role in shade avoidance response HD-Zip IIγ and HD-Zip IIδ proteins control several aspects of plant development, including embryo patterning, meristem function, leaf polarity and carpel development ().Citation28-Citation31
Figure 2. HAT3 and ATHB4 are required for vegetative and reproductive development in a white light environment. A) Wild-type (Col-0), hat3-3 athb4-128 and hat3-3 athb4-1 HAT3::HAT3:GFPCitation28 plants grown on soil for 5 wk in a L/D cycle (16/8 h) in white light. Bar: 1 cm. B) Siliques of Col-0 and hat3-3 athb4-128 plants grown as described in (A). White asterisks, junctions between peduncles and siliques. Bars: 1 mm. Growth conditions and light settings were as previously reported.Citation15

It has been in fact reported that cotyledon development and number are altered in hat3 athb4 embryos, and that these defects correlate with changes in auxin distribution and response, indicating a link between auxin-mediated embryo patterning and HD-Zip IIδ function.Citation28 Interestingly, it has also been shown that ATHB2 is induced in the HAT3-ATHB4 expression domain in the apical part of the embryo, marking the incipient cotyledons, as well as in the shoot apical meristem (SAM) of hat3 athb4, and thus can at least partially compensate for HAT3 and ATHB4 function.Citation28 HD-Zip IIγ and HD-Zip ΙΙδ proteins contain a LxLxL type of EAR repression motif,Citation7,Citation32 and there is evidence that at least some of them function as negative regulators of gene expression.Citation7,Citation15,Citation24,Citation33 Consistent with this, it has been demonstrated that ATHB2 is directly negatively regulated by HAT3 in vivo.Citation28 In agreement with ATHB2 being, to some extent, functionally interchangeable with HD-Zip IIδ proteins, it has been found that athb2 loss-of-function mutations enhance the hat3 athb4 phenotype. Indeed, it was observed that hat3 athb4 athb2 mutants display 1 or 2 radialized cotyledons and lack an active SAM, a phenotype reminiscent of that caused by loss-of-function mutations in the HD-Zip III genes PHABULOSA (PHB), PHAVOLUTA (PHV) and REVOLUTA (REV), master regulators of apical embryo development and primary determinants of adaxial cell fate.Citation28,Citation34-Citation36 It has been then demonstrated that simultaneous lack of both HAT3 and ATHB4 is associated with loss of adaxial identity of cotyledons and leaves whereas ectopic expression of HAT3, ATHB4 or ATHB2 results in upward leaf curling, a phenotype similar to that seen when HD-Zip III genes are expressed in both adaxial and abaxial regions of lateral organs ().Citation4,Citation28,Citation29,Citation37-Citation39 The functional interaction between HD-Zip II and HD-Zip III genes has been further corroborated by the discovery that ATHB2 is directly positively regulated by REV in vivo.Citation28
Figure 3. Ectopic expression of ATHB4 causes upward leaf curling. Wild-type (Ws) and 35S::ATHB4 (Ws) plants grown for 4 wk on soil in a L/D cycle (16/8h) in white light. The upward leaf curling phenotype was observed in several independent 35S::ATHB4 lines. White arrowheads, leaves displaying upward curling. Bars: 0.5 cm. Growth conditions and light settings were as previously reported.Citation15

The upstream region of all the HD-Zip II genes is enriched for HD-Zip binding sequences, and it has been shown that inducible chimeric proteins consisting of the DNA binding domain of HD-Zip II proteins and the transactivation domain of the herpes simplex virion protein 16 (VP16) directly induce the expression of all HD-Zip II genes in vivo, suggesting an intricate regulatory network within this gene family.Citation7,Citation24 Furthermore, there is evidence that the sites recognized in vitro by the HD-Zip II and HD-Zip III domains share the same core sequence (AAT[G/C]ATT).Citation40,Citation41 Consistent with this, chromatin immunoprecipitation followed by high-throughput sequencing using plants expressing a tagged ligand-binding domain of the glucocorticoid receptor, fused to a microRNA-resistant version of REV under control of the 35S-promoter has identified loci with the motif AT(G/C)AT, including the promoter regions of several HD-Zip II genes.Citation42 There is some recent evidence that closely-related sub-family members might share regulation of target genes through redundant promoter occupancy, in a way that differs from one gene to another.Citation43 It has been therefore suggested that the expression pattern of each HD-Zip II gene in a white light environment may depend on its responsiveness to the different HD-Zip III proteins, whose levels are tightly regulated both in time and space during development.Citation28,Citation44 The existence of a negative feedback network within the HD-Zip II family may add a further layer of complexity to the interactions among HD-Zip II and HD-Zip III proteins.Citation7,Citation28
HAT1 (also known as JAIBACitation30 [JAB]), ATHB2, HAT3 and ATHB4 have also been identified as genes positively regulated by SPATULA (SPT), a bHLH protein related to PIFs but lacking an active phytochrome binding domain involved in their negative regulation by the phytochrome in High R/FR, and proposed to be involved in carpel margin development.Citation31 SPT is known to play a key role in promoting growth of tissues derived from the carpel margins.Citation45 Indeed, loss of SPT function results in pronounced disruption of the septum and transmitting tract, decrease in stigmatic tissue development, and defects in apical carpel fusion.Citation46 The apical-basal patterning of the gynoecium appears to depend on the formation of an auxin gradient from the apex to the base, and several lines of evidence indicate close links between SPT function and auxin. For example, SPT has been proposed to promote and/or respond to the auxin peak that forms at the gynoecium apex.Citation47-Citation49 Consistent with a role for HD-Zip IIγ and HD-Zip IIδ genes as SPT targets in the development of carpel margin tissues, it has been found that loss-of-function hat3 athb4 double mutants have incomplete apical fusion in the gynoecium and severely reduced development of the septum as the spt mutants.Citation31 A pronounced gynoecium phenotype has also been observed in jab mutants, and there is evidence that JAB genetically interacts with CRABS CLAW (CRC), a member of the YABBY transcription factor family that is known to participate in the floral meristem (FM) determination process and in gynoecium development.Citation30,Citation45,Citation50,Citation51 It has been indeed found that jab crc double mutant flowers have an increase of organ number in 1 or more of the floral whorls and extra carpels inside the primary carpel, thus indicating that JAB, together with CRC, is involved in FM determination.Citation30 Moreover, it has been shown that jab combined with crc causes severe defects in gynoecium medial tissue development. In wild type the medial tissues grow toward each other and fuse in the center. In the jab mutant tissue fusion is similar to that of wild type while lack of fusion in the apical region of the gynoecia is evident in the crc mutant. In jab crc double mutant the medial tissues show reduced growth and do not fuse at all, indicating that JAB and CRC are both implicated in medial tissue proliferation.Citation30
In sum, recent work uncovered a crucial function for HD-Zip IIγ and HD-Zip IIδ genes in plant development from embryogenesis onwards. On one hand, it was found that HD-Zip II transcription factors control embryo apical patterning, SAM activity and organ polarity interacting with PHB, PHV and REV HD-Zip III proteins.Citation28,Citation29 On the other hand, JAB, ATHB2, HAT3 and ATHB4 were identified as genes positively regulated by the SPT bHLH transcription factor, and shown to participate in the regulation of carpel development.Citation30,Citation31 Interestingly, close links between auxin and both SPT and HD-Zip III function have been reported,Citation47-Citation49,Citation52 and a role for HAT3 and ATHB4 in promoting auxin responses during embryogenesis has emerged.Citation28
Most developmental auxin responses involve changes in gene expression, and depend on activation of AUXIN RESPONSE FACTORS (ARFs), transcription factors that positively or negatively regulate target genes.Citation53 ARFs are inhibited by AUXIN/INDOLE-3-ACETIC ACID (AUX/IAA) proteins, which are targeted for degradation by the 26S proteasome in response to auxin.Citation54,Citation55 For example, MONOPTEROS (MP)/ARF5 and its interacting Aux/IAA partner BODENLOS (BDL)/IAA12 play a central role in embryo patterning, and it has been shown that both loss-of-function mp mutants and gain-of-function bdl mutants exhibit a rootless phenotype and cotyledon defects.Citation56
There is compelling evidence that the action of auxin is context-dependent.Citation57,Citation58 For example, it has been recently reported that in the apical-most suspensor cell auxin promotes the specification of the hypophysis, the founder of the root apical meristem, and its subsequent asymmetric division whereas in the remaining suspensor cells auxin prevents proliferation.Citation59 How specificity is generated and how ARF and AUX/IAA gene family members contribute to local auxin response is not yet well understood. However, it has been demonstrated that the expression of the ARF genes is highly dynamic during development, and that distantly related ARF genes are often co-expressed. Interestingly, it has also been observed a strict correlation between cell type and the combination of ARFs expressed, suggesting that transcriptional regulation of ARF genes may underline the context-dependent action of auxin.Citation60 Furthermore, there is some evidence that, in addition to auxin-inducible degradation, Aux/IAAs are subject to spatio-temporal control of expression. It has been indeed recently found that the HD-Zip I transcription factor ATHB5 acts as a negative regulator of BDL expression, and thus might contribute to the exclusion of BDL from the epidermis and cortex.Citation61
The finding that an Aux/IAA gene is a direct target of a HD-Zip I family member is particularly interesting in light of the links between auxin and HD-Zip II proteins that have recently emerged. The 9 bp pseudopalyndromic sequences recognized in vitro with higher affinity by HD-Zip I and HD-Zip II proteins differ only at the central position (CAAT[A/T]ATTG and CAAT[G/C]ATTG, respectively).Citation8,Citation40 Furthermore, in vitro DNA binding studies and transient expression assays have demonstrated that ATHB1, a member of the HD-Zip I protein family, is able to recognize, although less efficiently, the sequence CAAT(G/C)ATTG. Reciprocally, ATHB2 recognizes the HD-Zip I binding site with a reduced affinity relative to the HD-Zip II one.Citation15,Citation62 Therefore, it is reasonable to hypothesize that HD-Zip IIγ and HD-Zip IIδ proteins may directly regulate BDL and potentially other Aux/IAA genes, contributing to spatio-temporal control of their expression. Given the complex role of HD-Zip IIγ and HD-Zip IIδ proteins in plant development and in shade avoidance, it will be a major challenge for future research to unravel their interactions with the auxin response machineries involved in patterning events and in growth responses to changes in the light environment.
Acknowledgments
We thank all our collaborators who made the work on HD-Zip II proteins a rewarding experience. Our apologies to the many researchers whose work or original publications could not be cited here because of space constraints. Authors work is funded by the Italian Ministry of Education, University and Research, FIRB-ERA-PG Program, the Italian Ministry of Agricultural, Food and Forestry Policies, NUTRIGEA Program, and the Italian Ministry of Economy and Finance, Project FaReBio di Qualità.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Ruberti I, Sessa G, Lucchetti S, Morelli G. A novel class of plant proteins containing a homeodomain with a closely linked leucine zipper motif. EMBO J 1991; 10:1787 - 91; PMID: 1675603
- Sessa G, Carabelli M, Ruberti I, Baima S, Lucchetti S, Morelli G. Identification of distinct families of HD-Zip proteins in Arabidopsis thaliana. In “Molecular-Genetic Analysis of Plant Development and Metabolism”, P Puigdomenech, G Coruzzi eds, Springer Verlag NATO-ASI series 1994; H 81:411-26.
- Morelli G, Baima S, Carabelli M, Di Cristina M, Lucchetti S, Sessa G, et al. Homeodomain-leucine zipper proteins in the control of plant growth and development. In “Cellular Integration of Signaling Pathways in Plant Development”, R Last, F Lo Schiavo, G Morelli, N Raikel eds, Springer Verlag NATO-ASI series 1998; H 104:251-62.
- Baima S, Possenti M, Matteucci A, Wisman E, Altamura MM, Ruberti I, et al. The arabidopsis ATHB-8 HD-zip protein acts as a differentiation-promoting transcription factor of the vascular meristems. Plant Physiol 2001; 126:643 - 55; http://dx.doi.org/10.1104/pp.126.2.643; PMID: 11402194
- Henriksson E, Olsson AS, Johannesson H, Johansson H, Hanson J, Engström P, et al. Homeodomain leucine zipper class I genes in Arabidopsis. Expression patterns and phylogenetic relationships. Plant Physiol 2005; 139:509 - 18; http://dx.doi.org/10.1104/pp.105.063461; PMID: 16055682
- Nakamura M, Katsumata H, Abe M, Yabe N, Komeda Y, Yamamoto KT, et al. Characterization of the class IV homeodomain-Leucine Zipper gene family in Arabidopsis. Plant Physiol 2006; 141:1363 - 75; http://dx.doi.org/10.1104/pp.106.077388; PMID: 16778018
- Ciarbelli AR, Ciolfi A, Salvucci S, Ruzza V, Possenti M, Carabelli M, et al. The Arabidopsis homeodomain-leucine zipper II gene family: diversity and redundancy. Plant Mol Biol 2008; 68:465 - 78; http://dx.doi.org/10.1007/s11103-008-9383-8; PMID: 18758690
- Ariel FD, Manavella PA, Dezar CA, Chan RL. The true story of the HD-Zip family. Trends Plant Sci 2007; 12:419 - 26; http://dx.doi.org/10.1016/j.tplants.2007.08.003; PMID: 17698401
- Schena M, Davis RW. HD-Zip proteins: members of an Arabidopsis homeodomain protein superfamily. Proc Natl Acad Sci USA 1992; 89:3894 - 8; http://dx.doi.org/10.1073/pnas.89.9.3894; PMID: 1349174
- Carabelli M, Sessa G, Baima S, Morelli G, Ruberti I. The Arabidopsis Athb-2 and -4 genes are strongly induced by far-red-rich light. Plant J 1993; 4:469 - 79; http://dx.doi.org/10.1046/j.1365-313X.1993.04030469.x; PMID: 8106086
- Carabelli M, Morelli G, Whitelam G, Ruberti I. Twilight-zone and canopy shade induction of the Athb-2 homeobox gene in green plants. Proc Natl Acad Sci USA 1996; 93:3530 - 5; http://dx.doi.org/10.1073/pnas.93.8.3530; PMID: 11607652
- Steindler C, Borello U, Carabelli M, Morelli G, Ruberti I. Phytochome A, phytochome B and other phytochome(s) regulate ATHB-2 gene expression in etiolated and green Arabidopsis plants. Plant Cell Environ 1997; 20:759 - 63; http://dx.doi.org/10.1046/j.1365-3040.1997.d01-123.x
- Franklin KA, Praekelt U, Stoddart WM, Billingham OE, Halliday KJ, Whitelam GC. Phytochromes B, D, and E act redundantly to control multiple physiological responses in Arabidopsis. Plant Physiol 2003; 131:1340 - 6; http://dx.doi.org/10.1104/pp.102.015487; PMID: 12644683
- Hornitschek P, Kohnen MV, Lorrain S, Rougemont J, Ljung K, López-Vidriero I, et al. Phytochrome interacting factors 4 and 5 control seedling growth in changing light conditions by directly controlling auxin signaling. Plant J 2012; 71:699 - 711; http://dx.doi.org/10.1111/j.1365-313X.2012.05033.x; PMID: 22536829
- Steindler C, Matteucci A, Sessa G, Weimar T, Ohgishi M, Aoyama T, et al. Shade avoidance responses are mediated by the ATHB-2 HD-zip protein, a negative regulator of gene expression. Development 1999; 126:4235 - 45; PMID: 10477292
- Morelli G, Ruberti I. Shade avoidance responses. Driving auxin along lateral routes. Plant Physiol 2000; 122:621 - 6; http://dx.doi.org/10.1104/pp.122.3.621; PMID: 10712524
- Morelli G, Ruberti I. Light and shade in the photocontrol of Arabidopsis growth. Trends Plant Sci 2002; 7:399 - 404; http://dx.doi.org/10.1016/S1360-1385(02)02314-2; PMID: 12234731
- Kanyuka K, Praekelt U, Franklin KA, Billingham OE, Hooley R, Whitelam GC, et al. Mutations in the huge Arabidopsis gene BIG affect a range of hormone and light responses. Plant J 2003; 35:57 - 70; http://dx.doi.org/10.1046/j.1365-313X.2003.01779.x; PMID: 12834402
- Carabelli M, Possenti M, Sessa G, Ciolfi A, Sassi M, Morelli G, et al. Canopy shade causes a rapid and transient arrest in leaf development through auxin-induced cytokinin oxidase activity. Genes Dev 2007; 21:1863 - 8; http://dx.doi.org/10.1101/gad.432607; PMID: 17671088
- Tao Y, Ferrer JL, Ljung K, Pojer F, Hong F, Long JA, et al. Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 2008; 133:164 - 76; http://dx.doi.org/10.1016/j.cell.2008.01.049; PMID: 18394996
- Keuskamp DH, Pollmann S, Voesenek LA, Peeters AJ, Pierik R. Auxin transport through PIN-FORMED 3 (PIN3) controls shade avoidance and fitness during competition. Proc Natl Acad Sci USA 2010; 107:22740 - 4; http://dx.doi.org/10.1073/pnas.1013457108; PMID: 21149713
- Li L, Ljung K, Breton G, Schmitz RJ, Pruneda-Paz J, Cowing-Zitron C, et al. Linking photoreceptor excitation to changes in plant architecture. Genes Dev 2012; 26:785 - 90; http://dx.doi.org/10.1101/gad.187849.112; PMID: 22508725
- Sassi M, Wang J, Ruberti I, Vernoux T, Xu J. Shedding light on auxin movement: Light-regulation of polar auxin transport in the photocontrol of plant development. Plant Signal Behav 2013; 8:e23355; http://dx.doi.org/10.4161/psb.23355; PMID: 23333970
- Sawa S, Ohgishi M, Goda H, Higuchi K, Shimada Y, Yoshida S, et al. The HAT2 gene, a member of the HD-Zip gene family, isolated as an auxin inducible gene by DNA microarray screening, affects auxin response in Arabidopsis. Plant J 2002; 32:1011 - 22; http://dx.doi.org/10.1046/j.1365-313X.2002.01488.x; PMID: 12492842
- Schena M, Lloyd AM, Davis RW. The HAT4 gene of Arabidopsis encodes a developmental regulator. Genes Dev 1993; 7:367 - 79; http://dx.doi.org/10.1101/gad.7.3.367; PMID: 8449400
- Sorin C, Salla-Martret M, Bou-Torrent J, Roig-Villanova I, Martínez-García JF. ATHB4, a regulator of shade avoidance, modulates hormone response in Arabidopsis seedlings. Plant J 2009; 59:266 - 77; http://dx.doi.org/10.1111/j.1365-313X.2009.03866.x; PMID: 19392702
- Ruberti I, Sessa G, Ciolfi A, Possenti M, Carabelli M, Morelli G. Plant adaptation to dynamically changing environment: the shade avoidance response. Biotechnol Adv 2012; 30:1047 - 58; http://dx.doi.org/10.1016/j.biotechadv.2011.08.014; PMID: 21888962
- Turchi L, Carabelli M, Ruzza V, Possenti M, Sassi M, Peñalosa A, et al. Arabidopsis HD-Zip II transcription factors control apical embryo development and meristem function. Development 2013; 140:2118 - 29; http://dx.doi.org/10.1242/dev.092833; PMID: 23578926
- Bou-Torrent J, Salla-Martret M, Brandt R, Musielak T, Palauqui JC, Martínez-García JF, et al. ATHB4 and HAT3, two class II HD-ZIP transcription factors, control leaf development in Arabidopsis. Plant Signal Behav 2012; 7:1382 - 7; http://dx.doi.org/10.4161/psb.21824; PMID: 22918502
- Zúñiga-Mayo VM, Marsch-Martínez N, de Folter S. JAIBA, a class-II HD-ZIP transcription factor involved in the regulation of meristematic activity, and important for correct gynoecium and fruit development in Arabidopsis. Plant J 2012; 71:314 - 26; http://dx.doi.org/10.1111/j.1365-313X.2012.04990.x; PMID: 22409594
- Reymond MC, Brunoud G, Chauvet A, Martínez-Garcia JF, Martin-Magniette ML, Monéger F, et al. A light-regulated genetic module was recruited to carpel development in Arabidopsis following a structural change to SPATULA. Plant Cell 2012; 24:2812 - 25; http://dx.doi.org/10.1105/tpc.112.097915; PMID: 22851763
- Kagale S, Links MG, Rozwadowski K. Genome-wide analysis of ethylene-responsive element binding factor-associated amphiphilic repression motif-containing transcriptional regulators in Arabidopsis. Plant Physiol 2010; 152:1109 - 34; http://dx.doi.org/10.1104/pp.109.151704; PMID: 20097792
- Ohgishi M, Oka A, Morelli G, Ruberti I, Aoyama T. Negative autoregulation of the Arabidopsis homeobox gene ATHB-2.. Plant J 2001; 25:389 - 98; http://dx.doi.org/10.1046/j.1365-313x.2001.00966.x; PMID: 11260495
- Emery JF, Floyd SK, Alvarez J, Eshed Y, Hawker NP, Izhaki A, et al. Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr Biol 2003; 13:1768 - 74; http://dx.doi.org/10.1016/j.cub.2003.09.035; PMID: 14561401
- Prigge MJ, Otsuga D, Alonso JM, Ecker JR, Drews GN, Clark SE. Class III homeodomain-leucine zipper gene family members have overlapping, antagonistic, and distinct roles in Arabidopsis development. Plant Cell 2005; 17:61 - 76; http://dx.doi.org/10.1105/tpc.104.026161; PMID: 15598805
- Smith ZR, Long JA. Control of Arabidopsis apical-basal embryo polarity by antagonistic transcription factors. Nature 2010; 464:423 - 6; http://dx.doi.org/10.1038/nature08843; PMID: 20190735
- Juarez MT, Kui JS, Thomas J, Heller BA, Timmermans MCP. microRNA-mediated repression of rolled leaf1 specifies maize leaf polarity. Nature 2004; 428:84 - 8; http://dx.doi.org/10.1038/nature02363; PMID: 14999285
- Mallory AC, Reinhart BJ, Jones-Rhoades MW, Tang G, Zamore PD, Barton MK, et al. MicroRNA control of PHABULOSA in leaf development: importance of pairing to the microRNA 5′ region. EMBO J 2004; 23:3356 - 64; http://dx.doi.org/10.1038/sj.emboj.7600340; PMID: 15282547
- Ochando I, Jover-Gil S, Ripoll JJ, Candela H, Vera A, Ponce MR, et al. Mutations in the microRNA complementarity site of the INCURVATA4 gene perturb meristem function and adaxialize lateral organs in arabidopsis. Plant Physiol 2006; 141:607 - 19; http://dx.doi.org/10.1104/pp.106.077149; PMID: 16617092
- Sessa G, Morelli G, Ruberti I. The Athb-1 and -2 HD-Zip domains homodimerize forming complexes of different DNA binding specificities. EMBO J 1993; 12:3507 - 17; PMID: 8253077
- Sessa G, Steindler C, Morelli G, Ruberti I. The Arabidopsis Athb-8, -9 and -14 genes are members of a small gene family coding for highly related HD-ZIP proteins. Plant Mol Biol 1998; 38:609 - 22; http://dx.doi.org/10.1023/A:1006016319613; PMID: 9747806
- Brandt R, Salla-Martret M, Bou-Torrent J, Musielak T, Stahl M, Lanz C, et al. Genome-wide binding-site analysis of REVOLUTA reveals a link between leaf patterning and light-mediated growth responses. Plant J 2012; 72:31 - 42; http://dx.doi.org/10.1111/j.1365-313X.2012.05049.x; PMID: 22578006
- Zhang Y, Mayba O, Pfeiffer A, Shi H, Tepperman JM, Speed TP, et al. A quartet of PIF bHLH factors provides a transcriptionally centered signaling hub that regulates seedling morphogenesis through differential expression-patterning of shared target genes in Arabidopsis. PLoS Genet 2013; 9:e1003244; http://dx.doi.org/10.1371/journal.pgen.1003244; PMID: 23382695
- Barton MK. Twenty years on: the inner workings of the shoot apical meristem, a developmental dynamo. Dev Biol 2010; 341:95 - 113; http://dx.doi.org/10.1016/j.ydbio.2009.11.029; PMID: 19961843
- Alvarez J, Smyth DR. CRABS CLAW and SPATULA, two Arabidopsis genes that control carpel development in parallel with AGAMOUS.. Development 1999; 126:2377 - 86; PMID: 10225997
- Alvarez J, Smyth DR. Crabs claw and Spatula genes regulate growth and pattern formation during gynoecium development in Arabidopsis thaliana.. Int J Plant Sci 2002; 163:17 - 41; http://dx.doi.org/10.1086/324178
- Nemhauser JL, Feldman LJ, Zambryski PC. Auxin and ETTIN in Arabidopsis gynoecium morphogenesis. Development 2000; 127:3877 - 88; PMID: 10952886
- Ståldal V, Sundberg E. The role of auxin in style development and apical-basal patterning of the Arabidopsis thaliana gynoecium. Plant Signal Behav 2009; 4:83 - 5; http://dx.doi.org/10.4161/psb.4.2.7538; PMID: 19649177
- Benková E, Michniewicz M, Sauer M, Teichmann T, Seifertová D, Jürgens G, et al. Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 2003; 115:591 - 602; http://dx.doi.org/10.1016/S0092-8674(03)00924-3; PMID: 14651850
- Bowman JL, Smyth DR. CRABS CLAW, a gene that regulates carpel and nectary development in Arabidopsis, encodes a novel protein with zinc finger and helix-loop-helix domains. Development 1999; 126:2387 - 96; PMID: 10225998
- Prunet N, Morel P, Thierry A-M, Eshed Y, Bowman JL, Negrutiu I, et al. REBELOTE, SQUINT, and ULTRAPETALA1 function redundantly in the temporal regulation of floral meristem termination in Arabidopsis thaliana. Plant Cell 2008; 20:901 - 19; http://dx.doi.org/10.1105/tpc.107.053306; PMID: 18441215
- Bowman JL, Floyd SK. Patterning and polarity in seed plant shoots. Annu Rev Plant Biol 2008; 59:67 - 88; http://dx.doi.org/10.1146/annurev.arplant.57.032905.105356; PMID: 18031217
- Guilfoyle TJ, Hagen G. Auxin response factors. Curr Opin Plant Biol 2007; 10:453 - 60; http://dx.doi.org/10.1016/j.pbi.2007.08.014; PMID: 17900969
- Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M. Auxin regulates SCF(TIR1)-dependent degradation of AUX/IAA proteins. Nature 2001; 414:271 - 6; http://dx.doi.org/10.1038/35104500; PMID: 11713520
- Santner A, Calderon-Villalobos LI, Estelle M. Plant hormones are versatile chemical regulators of plant growth. Nat Chem Biol 2009; 5:301 - 7; http://dx.doi.org/10.1038/nchembio.165; PMID: 19377456
- De Smet I, Lau S, Mayer U, Jürgens G. Embryogenesis - the humble beginnings of plant life. Plant J 2010; 61:959 - 70; http://dx.doi.org/10.1111/j.1365-313X.2010.04143.x; PMID: 20409270
- Kieffer M, Neve J, Kepinski S. Defining auxin response contexts in plant development. Curr Opin Plant Biol 2010; 13:12 - 20; http://dx.doi.org/10.1016/j.pbi.2009.10.006; PMID: 19942473
- Möller B, Weijers D. Auxin control of embryo patterning. Cold Spring Harb Perspect Biol 2009; 1:a001545; http://dx.doi.org/10.1101/cshperspect.a001545; PMID: 20066117
- Rademacher EH, Lokerse AS, Schlereth A, Llavata-Peris CI, Bayer M, Kientz M, et al. Different auxin response machineries control distinct cell fates in the early plant embryo. Dev Cell 2012; 22:211 - 22; http://dx.doi.org/10.1016/j.devcel.2011.10.026; PMID: 22264733
- Rademacher EH, Möller B, Lokerse AS, Llavata-Peris CI, van den Berg W, Weijers D. A cellular expression map of the Arabidopsis AUXIN RESPONSE FACTOR gene family. Plant J 2011; 68:597 - 606; http://dx.doi.org/10.1111/j.1365-313X.2011.04710.x; PMID: 21831209
- De Smet I, Lau S, Ehrismann JS, Axiotis I, Kolb M, Kientz M, et al. Transcriptional repression of BODENLOS by HD-ZIP transcription factor HB5 in Arabidopsis thaliana. J Exp Bot 2013; http://dx.doi.org/10.1093/jxb/ert137; PMID: 23682118
- Sessa G, Morelli G, Ruberti I. DNA-binding specificity of the homeodomain-leucine zipper domain. J Mol Biol 1997; 274:303 - 9; http://dx.doi.org/10.1006/jmbi.1997.1408; PMID: 9405140